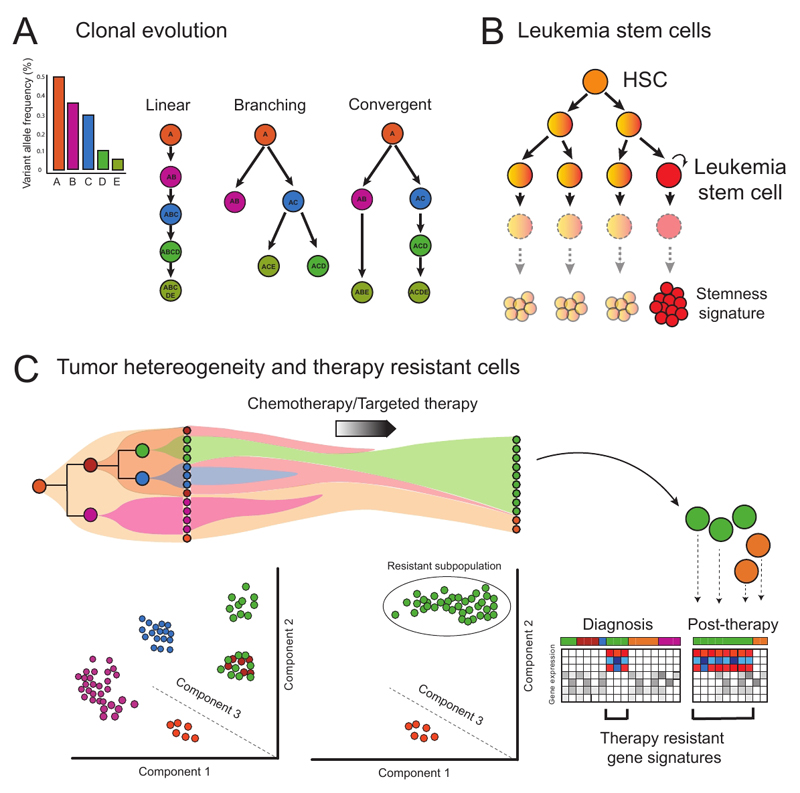
Figure 4. Concepts and applications of single cell genomics in resolving heterogeneity during malignant hematopoiesis.
(a) Single-cell sequencing is able to resolve different patterns of clonal evolution during tumour progression, providing valuable insights into pathways of cancer evolution. Each letter indicates a specific mutation. (b) Aberrant self-renewal in progenitor cell populations establishes leukemia stem cells. (c) Clonal dynamics and transcriptomic/epigenetic heterogeneity of leukemic populations can be revealed through single cell genomic techniques. The analysis of patient samples at diagnosis and after therapy provides crucial insights into different subpopulations and molecular pathways driving drug resistance and relapse. In the figure, a pre-leukemic population (orange) that acquires further mutations gives rise to a variety of leukemic subclones (purple, brown, green and blue). Single cell analysis can be used to identify these different subclones, importantly, also identifying additional heterogeneity within the green subclone, which is only uncovered by PCA (Principal Component Analysis). The pre-leukemic cells and one leukemia subclone (green) are resistant to therapy and the green subclone expands after treatment. The other subclones are eradicated, including a subpopulation of green cells. The molecular characterization of this persistent cell subpopulation can provide insights into mechanisms of therapy resistance that would not be obtained through analysis at the cell population level.