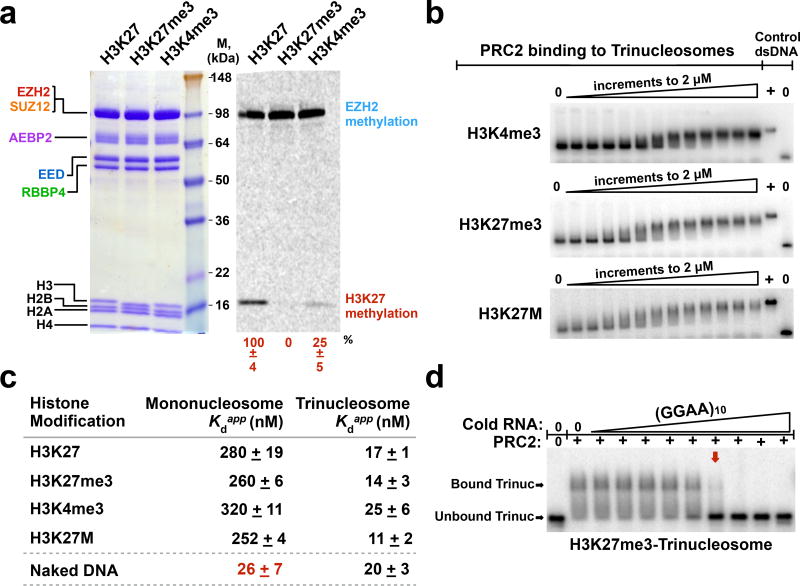
Figure 2.
Histone modifications have small effects on PRC2 affinity for nucleosome substrates in vitro. (a) Validation of unmodified and modified (H3K27me3 and H3K4me3) nucleosome variants using HMTase assays. PRC2 and histone proteins were visualized by Coomassie-staining (left-hand gel); methylation levels were visualized by 14C-autoradiography (right-hand gel). Quantification of methylation signal relative to unmodified nucleosomes +SD, n = 3 independent experiments. (b) Representative EMSA gels for PRC2 binding to 32P-radiolabeled trinucleosomes with indicated histone modifications. 2-fold titrations ranged from 0–2 µM. (c) Quantifications of PRC2 binding affinity to mono- and tri-nucleosomes containing different modifications. SD was calculated from three replicates. Bottom row shows Kd values for PRC2 binding to protein-free DNA controls (207 bp mononucleosome template DNA and 621 bp trinucleosome template DNA). (d) (GGAA)10 RNA disrupts pre-formed complexes of PRC2-H3K27me3-trinucleosome. PRC2-H3K27me3-trinucleosome complexes were fully disrupted (red arrow) at a PRC2:RNA molar ratio of ~2:1. Data points represent 3-fold titrations ranging from 0–10 µM RNA. Uncropped gel images are shown in Supplementary Data Set 1.