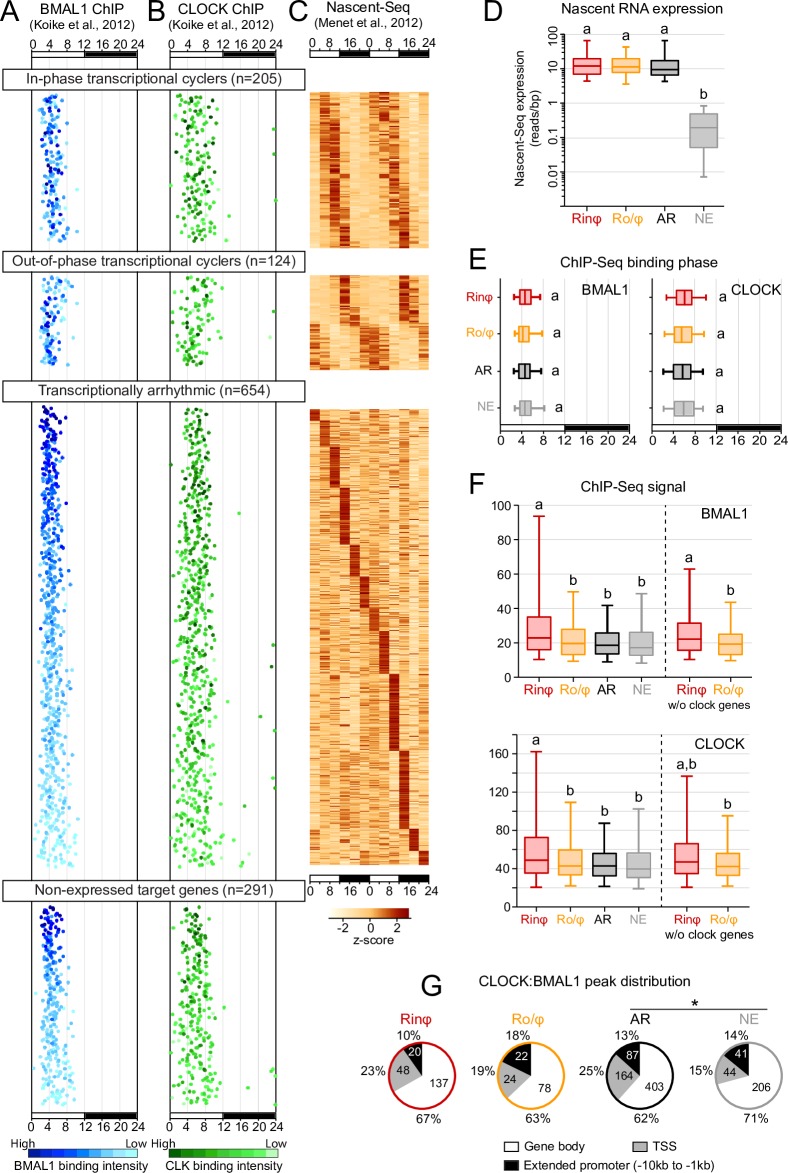
Fig 1. Mouse liver CLOCK:BMAL1 transcriptional output is heterogeneous.
A, B. Mouse liver BMAL1 (blue, A) and CLOCK (green, B) ChIP-Seq peaks from Koike et al., 2012 were mapped to their target genes and parsed based on their transcriptional output (Nascent-Seq from Menet et al., 2012, C). Each dot represents the phase of maximal DNA binding, and the ChIP-Seq signal is displayed using different shades of color to illustrate differences in binding intensity. C. Heatmap representation of the Nascent-Seq signal of direct CLOCK:BMAL1 target genes classified based on their transcriptional output in the mouse liver. Each lane represents the Nascent-Seq signal of a gene corresponding to the CLOCK and BMAL1 peaks in A and B. Nascent-Seq signal was ordered based on the phase of nascent RNA oscillations for the in-phase and out-of-phase transcriptional cyclers. Ordering of arrhythmically transcribed genes is based on the peak time of maximal expression; the lack of a distinctive 24-hr rhythm profile of nascent RNA expression over the 48-hr time-scale is indicative of arrhythmic transcription. No heatmap could be generated for the non-expressed genes because of the lack of nascent RNA expression. D. Nascent RNA expression, calculated as reads/bp, for each of the 4 CLOCK:BMAL1 transcriptional output group (Rinφ: in-phase transcriptional cyclers; Ro/φ: out-of-phase transcriptional cyclers; AR: arrhythmically transcribed target genes; NE: non-expressed target genes). Groups with different letters are significantly different (Kruskal-Wallis test; p < 0.05). E. Phase of maximal BMAL1 (left) and CLOCK (right) rhythmic DNA binding for each of the 4 CLOCK:BMAL1 transcriptional output categories. Groups with different letters are significantly different (Kruskal-Wallis test; p < 0.05). F. BMAL1 (top) and CLOCK (bottom) ChIP-Seq signal for each of the 4 CLOCK:BMAL1 transcriptional output groups. Signal is also displayed for the Rinφ and Ro/φ groups after removal of the ChIP-Seq signal at peaks targeting core clock genes. Groups with different letters are significantly different (Kruskal-Wallis test; p < 0.05). G. Location of CLOCK:BMAL1 peaks within gene loci for each of the 4 CLOCK:BMAL1 transcriptional output groups. TSS (transcriptional start site; +/- 1kb from annotated TSS); Gene body: + 1kb from TSS to + 1kb from transcription termination site; Extended promoter: - 10 kb to—1 kb from the annotated TSS. Numbers correspond to the percentage and numbers of peaks (outside and inside the pie chart, respectively) within each location for each group. * denotes a significant difference in the distribution of peaks between the AR and NE groups (chi square test; p < 0.05).