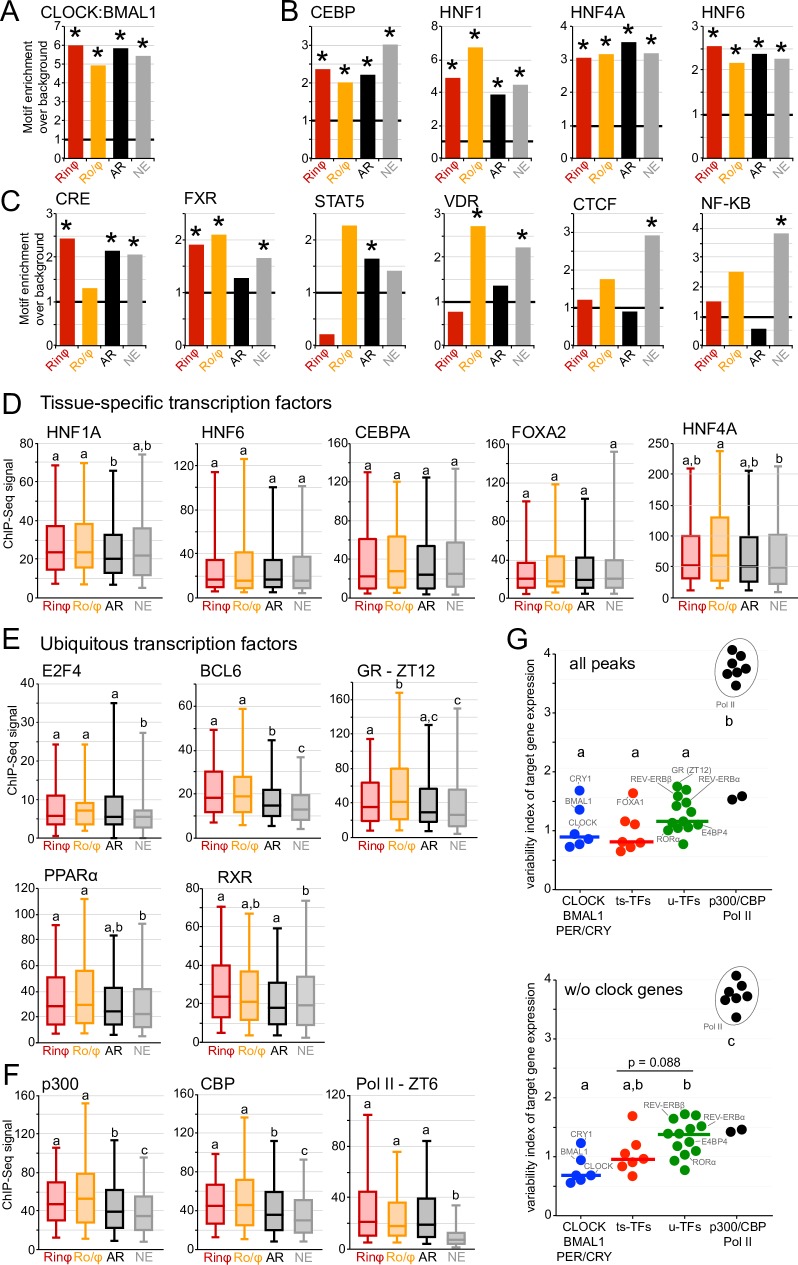
Fig 4. Tissue-specific and ubiquitous transcription factors are differentially recruited at CLOCK:BMAL1 enhancers.
A-C. Enrichment for the DNA binding motif of CLOCK:BMAL1 (A), tissue-specific transcription factors (B) and ubiquitous transcription factors (C) at CLOCK:BMAL1 DNA binding sites for each of the four CLOCK:BMAL1 transcriptional output categories. Enrichment was calculated using HOMER and is reported as the ratio between the calculated enrichment over the calculated background. * q < 0.05 (Benjamini-Hochberg procedure). D-F. ChIP-Seq signal of tissue-specific transcription factors (D), ubiquitous transcription factors (E), and transcriptional co-activators / RNA Polymerase II at ZT6 (F) at CLOCK:BMAL1 DNA binding sites (peak center ± 250bp) for each of the transcriptional output categories. Groups with different letters are significantly different (Kruskal-Wallis test; p < 0.05). G. Transcription factor DNA binding variability index at CLOCK:BMAL1 DNA binding sites. The TF DNA binding variability index reflects differential TF DNA binding by calculating the variance of TF ChIP-Seq signal between the four CLOCK:BMAL1 transcriptional output groups (see methods for details). The variability index is displayed as a dot for each TF: CLOCK, BMAL1, PER1, PER2, CRY1, CRY2 (blue), seven ts-TFs (CEBPA, CEBPB, FOXA1, FOXA2, HNF1, HNF4A, and HNF6; red), thirteen u-TFs (REV-ERBα, REV-ERVβ, RORα, E4BP4, RXR, LXR, PPARα, GR-ZT12, E2F4, STAT5, BCL6, ERα, and GABPA; green), as well as for p300, CBP, and Pol II at seven time points (ZT02 to ZT26) (black). The horizontal lines represent the variability index median for the first 3 groups of TF. ChIP-Seq datasets used in this analysis are described in the method section. The variability index was calculated using all CLOCK:BMAL1 peaks analyzed in Fig 1 (left), or CLOCK:BMAL1 peaks that do not target a clock gene (removal of TF ChIP-Seq signal at peaks targeting Per1, Per2, Cry2, Dbp, Rev-erbα, and Rev-erbβ, Tef, Hlf, Gm129, and Rorγ; right).