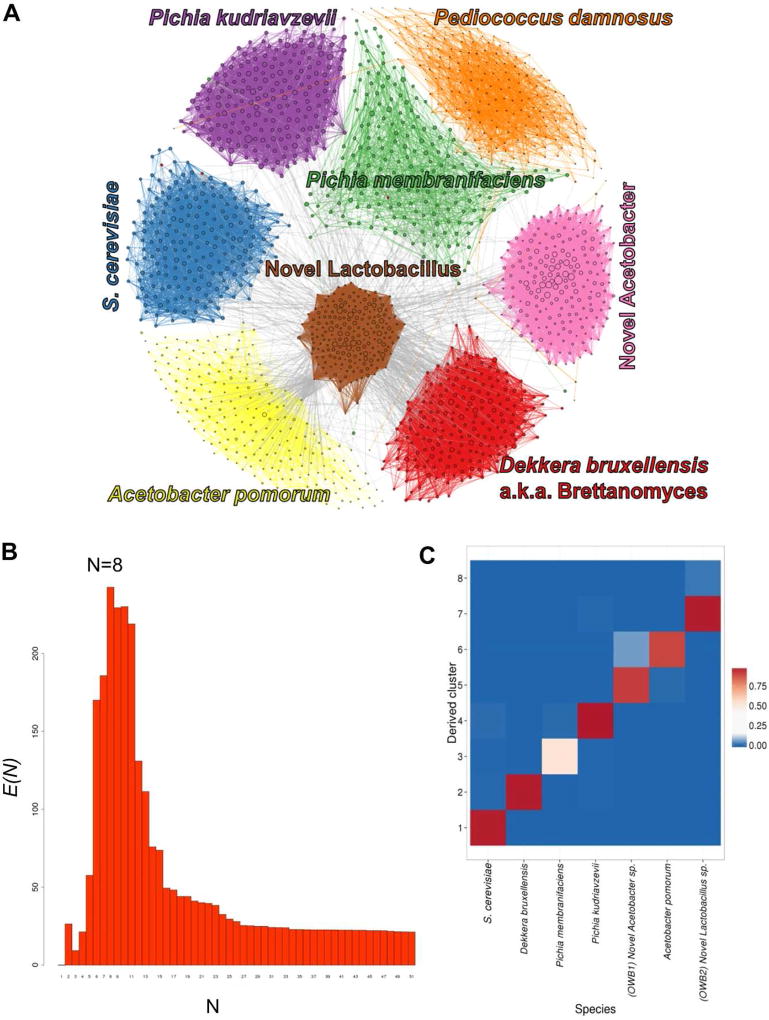
Figure 1. MetaPhase deconvolutes species from a spontaneously inoculated beer sample.
A. Clustering of the Old Warehouse assembly. Each contig is shown as a dot, with size indicating contig length, colored by species. Edge widths represent the densities of Hi-C links between the contigs shown. B. Determining the approximate number of species in the metagenome. We ran the hierarchical agglomerative clustering algorithm on the draft assembly. In this algorithm, the number of clusters starts high and gradually decreases as clusters are merged together; to generate this data, we continued clustering all the way down to N = 1 cluster. Shown is the metric E, or intra-cluster link enrichment, at each value of N. It is assumed that the maximum value of E(N) occurs when N is roughly equal to the true number of distinct species present in the draft assembly. For this assembly, this occurs at N = 8 clusters. C. Validation. This heatmap indicates what fraction of the sequence in each MetaPhase cluster maps uniquely to each of the reference genomes of the predicted species. Note that not all sequence is expected to map uniquely to one species. x-axis: the 7 species. y-axis: the MetaPhase clusters. Note, this excludes the P. damnosus-like cluster because there was no public P. damnosus reference genome available.