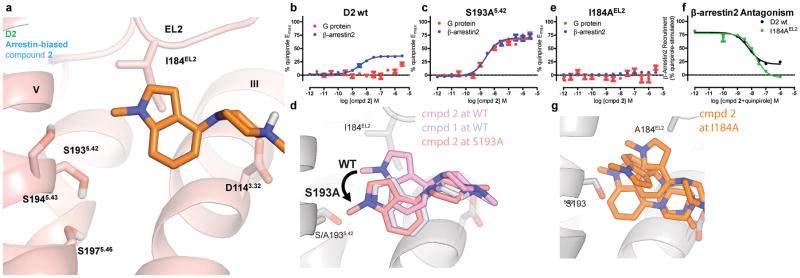
Figure 4. D2 TM5 and EL2 Mutants Confirm Arrestin-Bias Binding Pose.
a) The pose resulting from MD simulation of the head group of arrestin-biased N-methyl indole-aripiprazole hybrid (2) places the N-methyl indole moiety in contact with I184 on EL2, having moved away from S193 on TM5. b) N-methyl indole-aripiprazole hybrid 2 only shows arrestin recruitment activity at D2 wild-type. Data represent mean and standard error of the mean performed in triplicate (Gi/o GloSensor; red, n=3) and β-arrestin2 recruitment (Tango; blue, n=3, EC50 = 3.7 nM, Emax = 36%) c) S193A5.42 transforms arrestin-bias of 2 into balanced signaling with respect to quinpirole. Data represent Gαi/o-mediated cAMP inhibition (Gi/o GloSensor; red, n=3, EC50 = 2.5 nM, Emax = 67%) and β-arrestin2 recruitment (Tango; blue, n=3, EC50 = 2.6 nM, Emax = 69%). d) Representative pose of compound 2 head group from simulation at wild-type (WT) and S193A D2R constructs, and compound 1 head group from wild-type D2R simulation. At S193A, 2 moves to a pose almost identical to 1 at D2 wild-type. e) Mutation of EL2 I184 (I184A) completely abolishes arrestin recruitment for arrestin-biased ligand 2 (Tango; n=5 in triplicate) f) I184A mutation transforms 2 into a D2R β-arrestin2 recruitment antagonist as measured in Tango (n=2, in triplicate), as seen by comparing D2 wild-type (black, IC50 = 6.3 nM) to EL2 I184A (green, IC50 = 13 nM). g) Compound 2 head group is unstable throughout simulation at I184A D2R, sampling many orientations within the ligand-binding pocket. Ligand poses are shown for three points in time during a single simulation.