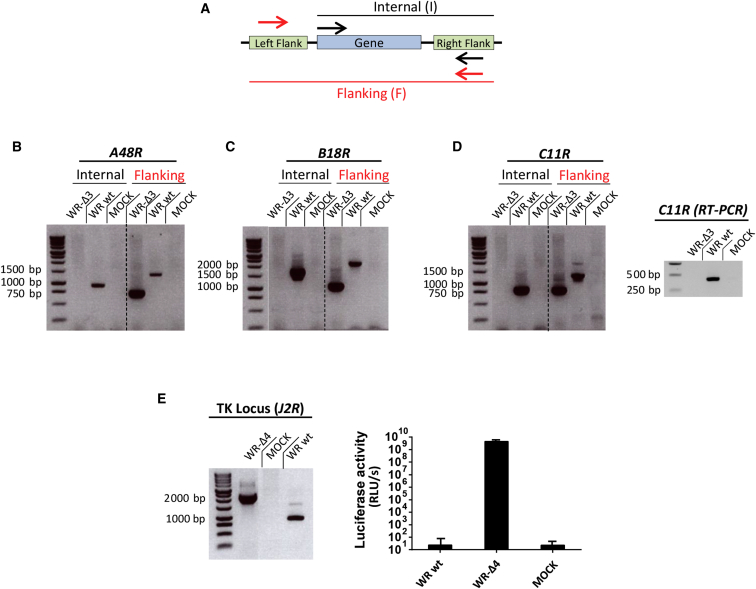
Figure 1.
Generation of the WR-Δ3 and WR-Δ4 Viruses
Viral genes were deleted sequentially from the WR parental virus through homologous recombination using deletion plasmids. (A) PCR strategy was used to confirm the mutant viruses; arrows indicate the amplification regions termed “flanking” (red) and “internal” (black). (B–D) PCR analyses using internal and flanking primer sets were used to confirm deletion of (B) A48R (internal: WR WT, 1,031 bp; flanking: WR WT, 1,423 bp, WR-Δ3, 740 bp), (C) B18R (internal: WR WT, 1,469 bp; flanking: WR WT, 1,890 bp, WR-Δ3, 835 bp), and (D) C11R (internal: WR WT, 794 bp; flanking: WR WT, 1,172 bp, WR-Δ3, 750 bp; RT-PCR: WR WT, 422 bp). To delete the J2R gene, we used homologous recombination to replace the viral gene with the luciferase reporter gene. (E) Left: PCR with flanking primers was used to confirm J2R deletion/luciferase insertion (WR WT, 843 bp; WR-Δ4, 2,106 bp); right: luciferase activity was assayed using a luminometer. Data are shown as mean ± SD. Samples were obtained from infected BSC-40 cells (MOI 1 PFU/cell).