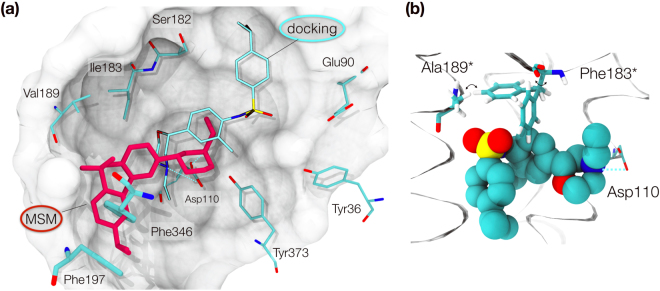
Figure 5.
Comparison of docking and MSM poses and effect of mutations. (a) Overlay of the pose obtained by high-throughput molecular dynamics (red), the docking prediction (blue) and the residues implicated in binding in the 3PBL coordinates. The new pose obtained by MSM fills the pocket formed by V1895.39A, and I183ECL2F and would clash with F1975.47and F3466.52 in their crystal structure coordinates. (b) The pose obtained by molecular dynamics provides a rationale for the effect of the two mutations (V1895.39A, I183ECL2F). The mutated residues are shown with an asterisk. Rotation of the engineered F183ECL2 would clash with either the ligand or residue 189, affecting protein stability and ligand affinity. Mutant A1895.39 would lose favorable packing (van der Waals) interactions by reduction of its sidechain.