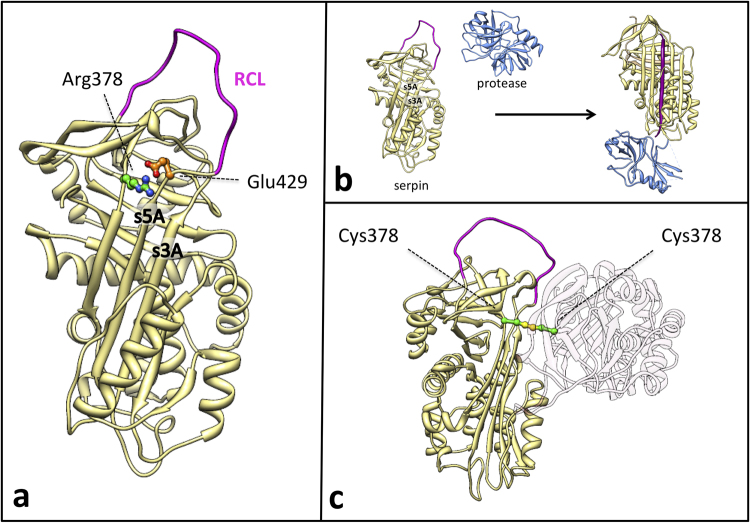
Figure 1.
(a) Structural localization of the Arg378Cys missense variants in the C1-INH serpin domain. Active inhibitory serpins in their native state are characterized by a central β-sheet (β-sheet A) and an exposed Reactive Centre Loop (RCL in purple) presenting the bite for the target protease. Arg378 (depicted as ball & stick, with C atoms in green and N atoms in blue), at the top of the β-sheet A, is expected to form a salt bridge with Glu429 (depicted as ball & stick, with C atoms in orange and O atoms in red) in the hinge of the RCL, that may regulate the opening of the β-sheet A at the insertion of the RCL between strand 3 (s3A) and strand 5 (s5A). (b) Serpins inhibitory mechanism. After the docking of the free protease (blue) and the active serpin (khaki), the protease cleaves the RCL (purple). RCL cleavage triggers a profound conformational change within the serpin molecule: the RCL downstream of the scissile bond inserts between strands 3 and 5 of β-sheet A as strand 4 while the protease is translocated and crushed against the opposite pole of the inhibitor molecule, resulting in enzyme inhibition by active site distortion. This conformational rearrangement greatly augments the stability of the serpin molecule. Insertion of the RCL in the structural core of the serpin can also happen without RCL cleavage (latentisation or polymerisation process) (c) Model of the disulfide-linked dimer of mutated C1-INH. Two mutated C1-INH molecules (khaki and light pink) interact in a dimer. The two Cys at position 378 (depicted as ball & stick, with C atoms in green and S atoms in yellow) form a disulphide bridge (highlighted in yellow) that covalently bounds the two molecules. The second molecule has been drawn nearly transparent for clarity purposes. We must consider that the disulphide bridge does not necessarily occur between native-like structures as reported in the picture. The coordinates used are from pdb entry 1M6Q for C1-INH as active serpin (homology model of the native C1-INH serpin domain), 1K1L for free protease, and 1EZY for serpin-protease complex. Figures were produced using Chimera72.