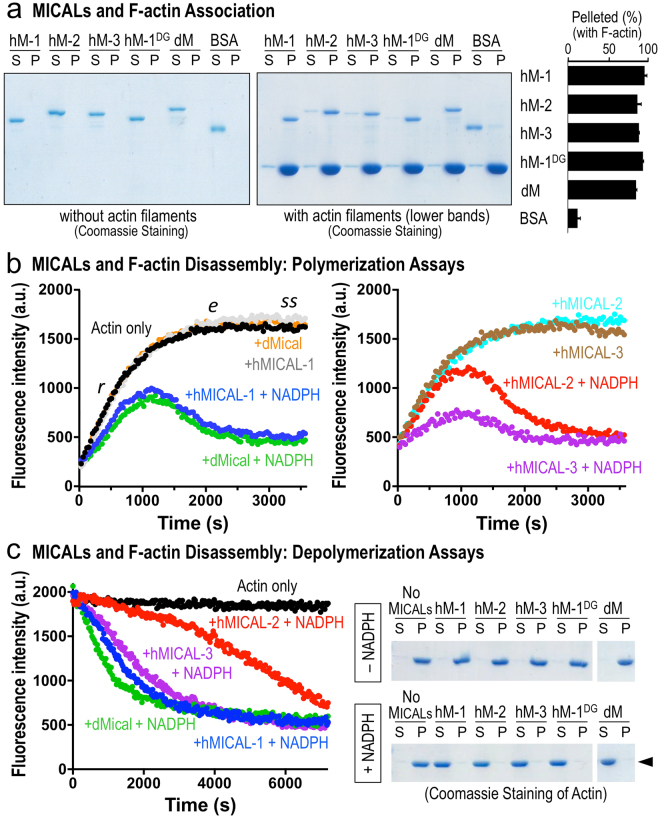
Figure 2.
Each human MICAL family member directly binds and disassembles actin filaments. (a) Each of the purified human MICALredoxCH proteins, hMICAL-1 (hM-1), hMICAL-2 (hM-2), hMICAL-3 (hM-3), and hMICAL-1 DG (hM-1DG), like Drosophila MicalredoxCH (dM), associates with F-actin as revealed by actin co-sedimentation/pelleting assays in which Coomassie blue stained gels are shown. Notice that after high-speed centrifugation, each of the purified MICALs is present in the soluble (S) fraction (left gel). In the presence of purified actin filaments, however, each of the MICALs is present in the pellet (P) fraction (right gel). The percentage (±the standard error of the mean (SEM)) of different MICAL proteins in the pelleted fraction following incubation with purified F-actin was quantified by densitometry (n = 3). Bovine serum albumin (BSA) was used as a control and was predominantly found in the S fraction. [MICALs] = 600 nM, [BSA] = 600 nM, [actin] = 2.3 μM. (b,c) Each of the human MICALs alters actin polymerization and depolymerization. Pyrene-labeled actin was used to monitor both the polymerization and depolymerization of actin using standard approaches, where the fluorescence intensity (a.u. (arbitrary units)) of the pyrene-labeled actin polymer is substantially higher than the pyrene-labeled actin monomer. Each of the MICALs and the reaction conditions are color-coded here and in Figs 3,5. (b) Standard pyrene-actin polymerization assay. No filaments are present at Time = 0, when MICALs are added to actin and polymerization is induced. As can be observed by following the characteristic increase in fluorescence intensity over time, the addition of each of the purified human MICALs (in the presence of their co-enzyme NADPH) decreases the rate (r), extent (e), and steady-state level (ss) of actin polymerization (as compared to an actin only control, black dots; or MICAL only (no NADPH) + actin controls; or NADPH only + actin controls (not shown; see11)). Notably, MICAL (NADPH) induces actin polymerization to slow-down over time, which is followed by a substantial decrease in the extent of polymerization, the rapid depolymerization of F-actin, and the inability of actin to reinitiate polymer formation. Thus, because each of the MICALs acts on F-actin11,14 (and herein), our data in the polymerization assays are consistent with a model in which as the actin begins to polymerize, the MICALs oxidize individual subunits of the polymer, which disassembles the polymerizing actin. This slows polymerization – and also creates fewer and fewer unoxidized monomers that can be used for polymerization. [MICAL-1] = 300 nM, [dMical and MICALs-2, 3] = 600 nM, [NADPH] = 100 μM, [Actin] = 1.15 μM. (c) Purified human MICALs+/– NADPH was added to actin that was polymerized and kept in buffer conditions that favored polymerization (as can be seen with the steady-state fluorescence intensity level of the actin only control (black dots); or actin with each of the MICALs without NADPH (not shown; see11)). The addition of each of the human MICALs (in the presence of NADPH) induces actin depolymerization (decreasing fluorescence intensity that can be followed over time in the pyrene actin depolymerization assay (left)). [MICAL-1] = 300 nM, [dMical and MICALs-2, 3] = 600 nM, [NADPH] = 100 μM, [Actin] = 1.15 μM. The extent of this MICAL (NADPH)-dependent actin depolymerization is also observed when MICAL-treated F-actin was subjected to high-speed centrifugation to differentiate F-actin (P) from G-actin (S). As can be observed in these Coomassie stained gels, each of the human MICALs (in the presence of NADPH) substantially increases the ratio of G-actin to F-actin (arrowhead). [MICAL-1] = 300 nM, [dMical and MICALs-2, 3] = 600 nM, [NADPH] = 100 μM, [Actin] = 1.15 μM. Unprocessed original scans of gels are shown in Supplementary Fig. 11.