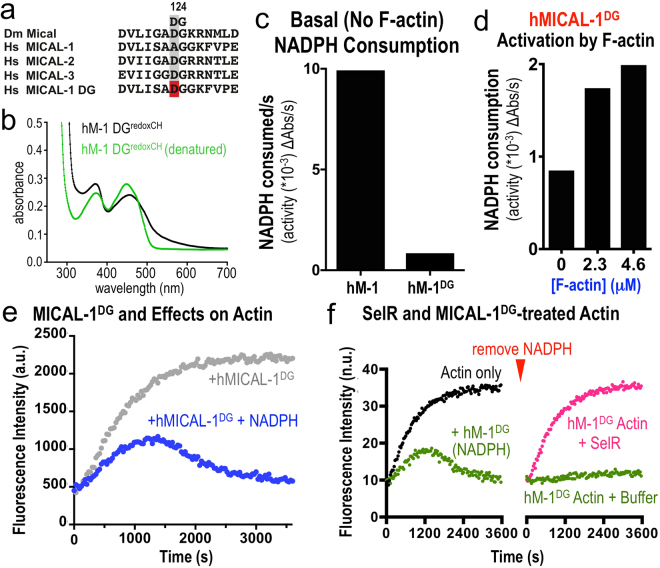
Figure 5.
MICAL-1 exhibits a single amino acid alteration in its DG motif that produces high levels of catalytic activity in the absence of its F-actin substrate. (a) MICAL-1 has a “naturally” occurring substitution of an alanine (A) residue instead of the important aspartate (D) residue in the DG (Conserved) motif that is present in the other MICAL family members. Using site-directed mutagenesis we converted the alanine residue within the DG motif of MICAL-1 to an aspartate residue and thereby generated a MICAL-1 protein similar to the other MICAL family members (MICAL-1DG). (b) Purified MICAL-1DG protein still exhibits the hallmarks of an FAD-binding protein. In particular, MICAL-1DG maintains its UV-visible light absorption spectra (with peaks at ~360 nm and ~450 nm, black lines), and denaturation of the MICAL-1DG releases FAD, which underlies this absorption spectra (green line). [MICAL-1DG] = 20 μM. (c) Converting the alanine residue within the DG motif of MICAL-1 to an aspartate residue (MICAL-1DG), substantially reduces (12-fold per second) the basal (in the absence of its F-actin substrate) enzyme activity of MICAL-1, as judged by the consumption of NADPH. [MICALs] = 600 nM, [NADPH] = 200 μM. (d) The enzymatic activity of MICAL-1DG, similar to other MICALs, is notably increased in the presence of F-actin. Enzyme activity was determined by the consumption (conversion of NADPH to NADP+) of MICAL’s co-enzyme NADPH, which was monitored by recording the light absorbance at 340 nm wavelength/time. [MICAL-1DG] = 600 nM, [NADPH] = 200 μM. (e) MICAL-1DG, similar to unaltered MICAL-1 and other MICAL family members, induces actin polymerization to slow-down over time, which is followed by a substantial decrease in the extent of polymerization, the rapid depolymerization of F-actin, and the inability of actin to reinitiate polymer formation. [Actin] = 1.15 μM, [MICAL-1DG] = 600 nM, [NADPH] = 100 μM. Here, as in Fig. 2b, pyrene-labeled actin was used to monitor both the polymerization and depolymerization of actin using standard approaches, where the fluorescence intensity (a.u. (arbitrary units)) of the pyrene-labeled actin polymer is substantially higher than the pyrene-labeled actin monomer. (f) Similar to unaltered MICAL-1 and other MICAL family members, the stereospecific methionine sulfoxide reductase SelR/MsrB restores the polymerization properties of actin treated with MICAL-1DG. See also Fig. 3d. Buffer (buffer that SelR is stored in), n.u. (normalized units between the 2 graphs). [Actin] = 1.15 μM, [MICAL-1DG] = 600 nM, [NADPH] = 100 μM.