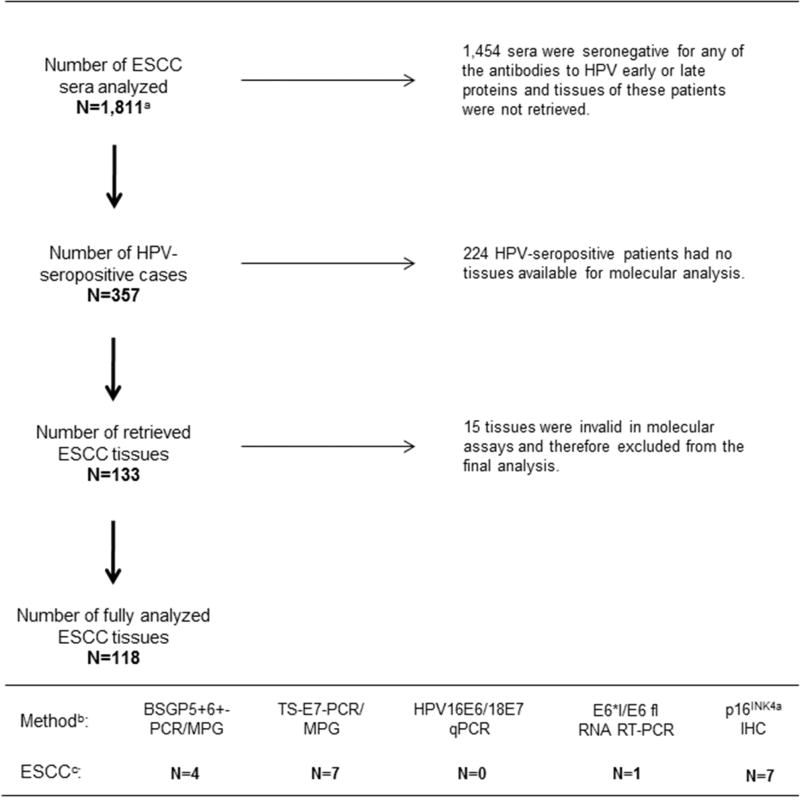
Figure 1. Overview of serological and tissue analyses in InterSCOPE study.
a 1,565 of the 1,811 sera had sufficient covariate data that were included in a previous case-control analysis (Sitas et al., 2012). b 118 ESCC tissues were analyzed using five molecular methods to assess: (i) presence of HPV DNA (BSGP5+6+-PCR/MPG and TS-E7-PCR/MPG), (ii) viral load (HPV16/18 E6 qRT-PCR), (iii) HPV mRNA (E6*I/E6 fl RNA RT-PCR), and (iv) expression of p16INK4a protein as a surrogate marker of HPV-transformed phenotype (p16INK4a IHC). c Of total 118 ESCC tissues, 10 (8%) were HPV DNA+ when results of both genotyping methods were combined, none of the HPV16 DNA+ tissues showed high viral load, and only a single HPV16 DNA+ case expressed viral mRNA (HPV16 DNA+/mRNA+/p16INK4a−). Seven ESCC tissues showed up-regulation of p16INK4a and only one tissue with p16INK4a up-regulation was HPV16 DNA+ but showed no positivity for HPV16 mRNA (HPV16 DNA+/mRNA−/p16INK4a +).