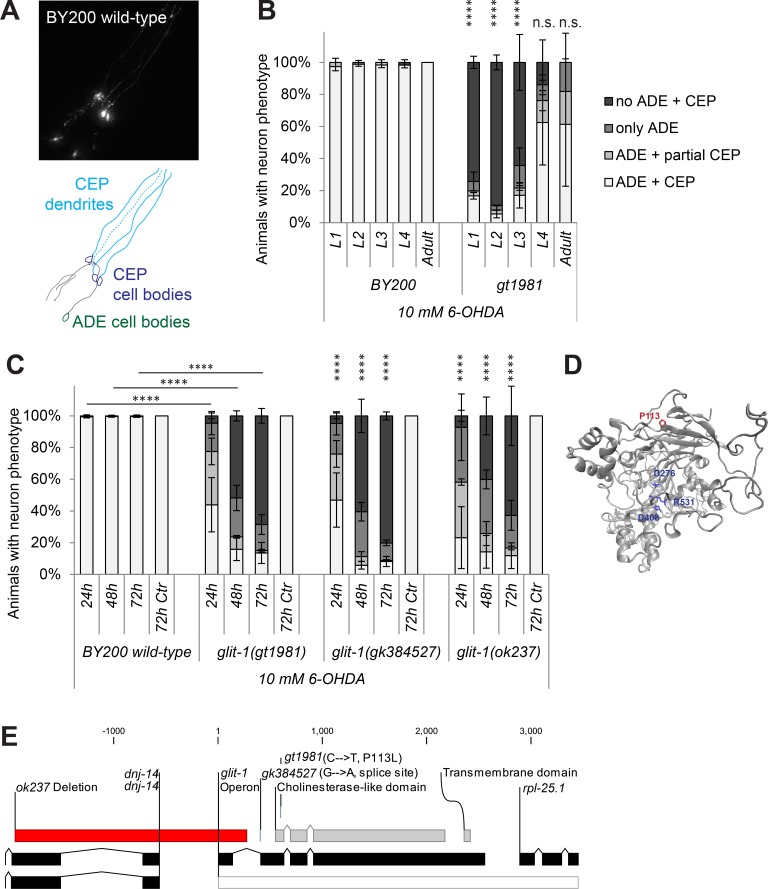
Fig 1. glit-1(gt1981) exhibits increased 6-OHDA-induced dopaminergic neurodegeneration.
(A) GFP-labelled C. elegans dopaminergic head neurons– 4 CEP neurons and 2 ADE neurons–in BY200 wild-type animals. (B) Remaining dopaminergic head neurons in BY200 wild-type animals and gt1981 mutants 48 hours after treating L1-L4 larval stages or adult animals with 10 mM 6-OHDA. Animals possessing all neurons were scored as ‘ADE + CEP’ (white bar), those with partial loss of CEP but intact ADE neurons as ‘ADE + partial CEP’ (light grey bar), those with complete loss of CEP but intact ADE neurons as ‘only ADE’ (dark grey bar) and those with complete loss of dopaminergic head neurons as ‘no ADE + CEP’ (black bar). Error bars = SEM of 2 biological replicates, each with 25–40 animals per stage and strain. Total number of animals per condition n = 50–80 (****p<0.0001, n.s. p>0.05; G-Test comparing BY200 wild-type and mutant data of the same lifecycle stages). (C) Dopaminergic head neurons 24, 48 and 72 hours after treatment with 10 mM 6-OHDA and 72 hours after treatment with ascorbic acid only (‘72h Ctr’) for BY200 wild-type or glit-1 mutant animals. Error bars = S.E.M. of 2 biological replicates for glit-1(ok237) and 3 biological replicates for all the other strains, each with 60–115 animals per strain and concentration. Total number of animals for the ‘72h Ctr’ experiment n = 30–100 and for all other conditions n = 130–340 (****p<0.0001; G-Test comparing BY200 wild-type and mutant data of the same time point). (D) GLIT-1 protein structure prediction based on homology modelling with acetylcholinesterase (PDB ID: 2W6C). The gt1981 point mutation leads to a proline to glycine conversion (P113G) and is indicated in red. The amino acids replacing the acetylcholinesterase catalytic triad are indicated in blue. (E) glit-1 gene structure with positions of the glit-1(gt1981) point mutation, the glit-1(gk384527) splice site mutation and the glit-1(ok237) deletion. For the point mutations the nucleotide and amino acid changes are indicated in brackets. The ok237 deletion spans 5’UTR and first exon of glit-1 and 5’UTR and first exons of dnj-14 (DNaJ domain (prokaryotic heat shock protein)) and is indicated with a red bar. Also, glit-1 is located in an operon (grey bar) with the ribosomal protein rpl-25.1 (Ribosomal Protein, Large subunit).