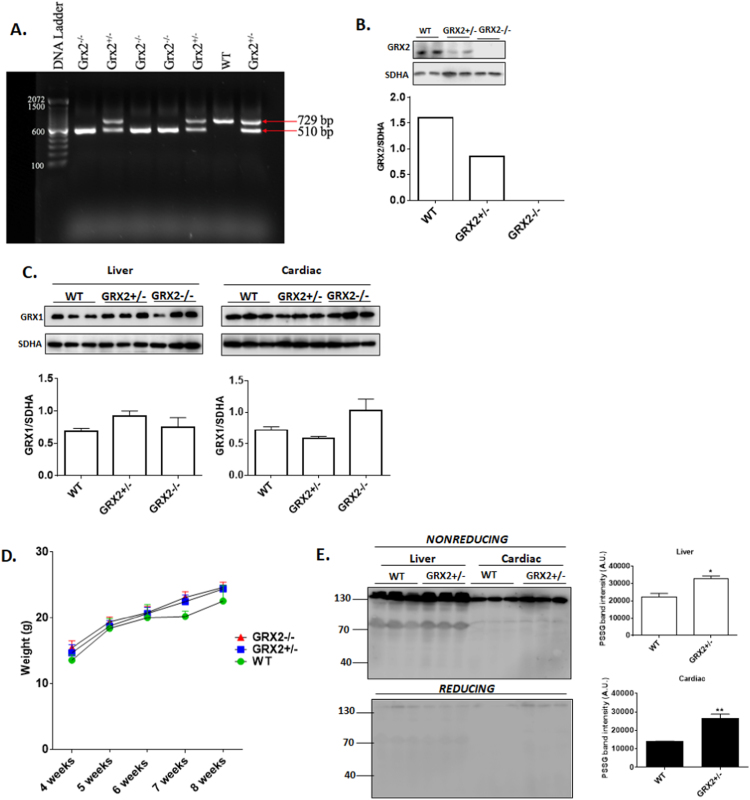
Fig. 2.
Impact of GRX2 deletion on the mitochondrial S-glutathionylated proteome and the expression of GRX1. A. Genotypic characterization of mouse litters. RNA was extracted from ear notches and subjected to PCR analysis for the presence of the full length or truncated forms of the Grx2 gene. B. Immunoblot analysis confirming the partial knockout and complete abolishment of GRX2. Assessment of GRX2 protein levels was conducted on mitochondria isolated from liver tissue. SDH subunit A served as the loading control. C. Depletion or abolishment of GRX2 does not induce a compensatory increase in GRX1 in cardiac or liver tissue. Band intensities were quantified by ImageJ. SDH subunit A served as the loading control. n = 3, mean ± SEM. 1-way ANOVA Fisher's LSD post-hoc test. D. GRX2+/- and GRX2-/- do not display a significant change in overall body weight when compared to WT littermates. n = 12–21, mean ± SEM, 1-way ANOVA Fisher's LSD post-hoc test. E. Partial ablation of the Grx2 gene induces a significant increase in the overall number of S-glutathionylated proteins in cardiac and liver mitochondria. Samples were electrophoresed under reducing (+ 2% v/v β-mercaptoethanol) or nonreducing (no β-mercaptoethanol) and probed for the presence of S-glutathionylated proteins using protein-glutathione disulfide (PSSG) anti-serum. n = 3, mean ± SEM. 1-way ANOVA Fisher's LSD post-hoc test.