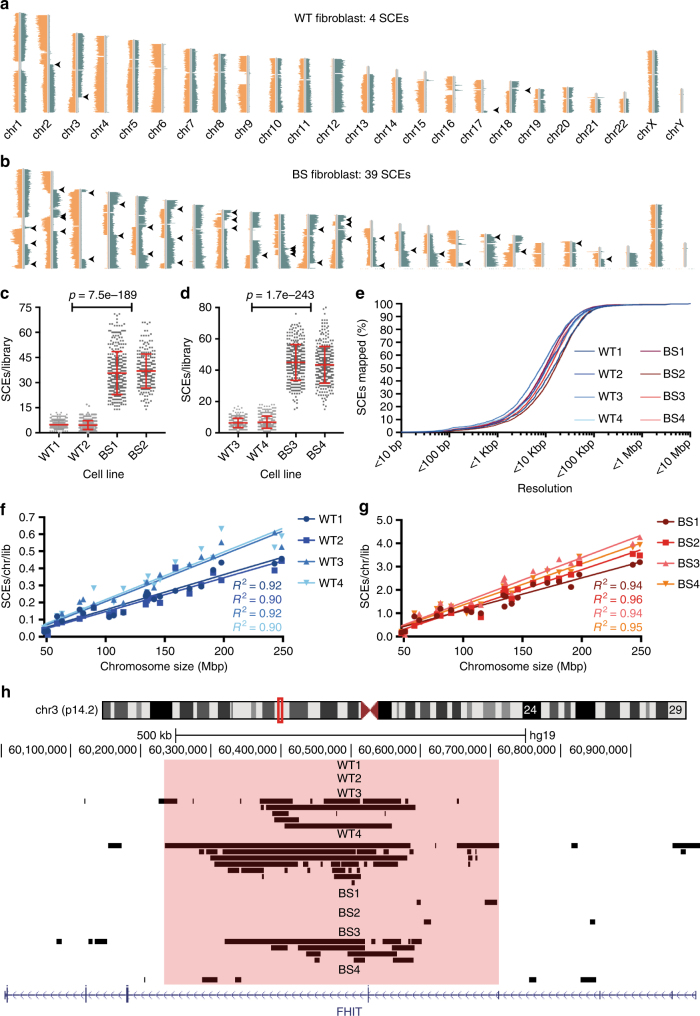
Fig. 1.
High-resolution mapping of SCEs and common fragile site hotspots. a, b Representative Strand-seq libraries generated from a a WT fibroblast and b a BS fibroblast. Mapped DNA template strand reads are plotted on directional chromosome ideograms; reads mapping to the Crick (positive) strand of the reference genome are shown in green, those mapping to the Watson (negative) strand are shown in orange. SCEs are identified as a switch in template strand state, indicated by arrowheads. c, d Number of SCEs detected during a single cell cycle in c primary fibroblasts and d EBV-transformed B-lymphocytes obtained from healthy donor and BS patients. Each grey point represents number of SCEs detected in a single-cell Strand-seq library, red lines indicate mean ± SD. p-values were calculated using ANOVA. e SCE mapping resolutions across all eight cell lines. Lines represent percentage of the total number of SCEs mapped at resolutions below indicated values. f, g Correlations between average numbers of SCEs/chromosome/library and chromosome size for f WT and g BS cells. R2-values are color-matched to the cell lines. h Example of SCE hotspot detected within FRA3B (FHIT). Mapped SCE regions for each cell line were uploaded onto the UCSC Genome Browser. Black bars represent genomic locations of SCE regions; size indicates mapping resolution using the BAIT program. Red box indicates the location of the SCE hotspot as detected by Strand-seq