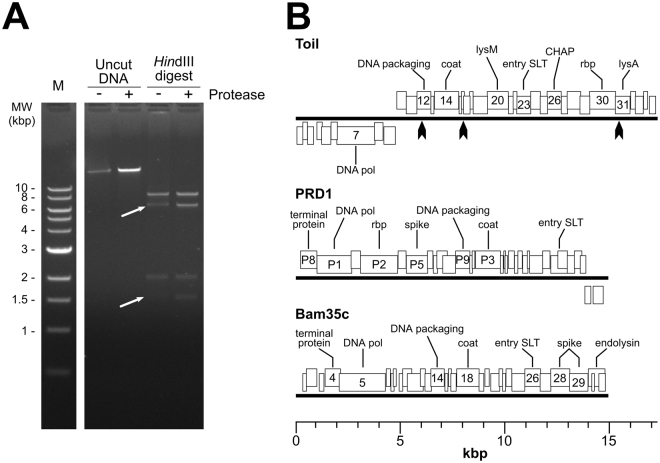
Figure 2.
Genome analysis of phage Toil. (A) Restriction digest of phage Toil genomic DNA with and without proteinase K treatment. Lane 1, NEB 1 kb molecular weight standard; Lane 2, Untreated DNA; Lane 3, proteinase K treated DNA; Lane 4, HindIII digested DNA; Lane 5, HindIII digested and proteinase treated. White arrows in lane 4 point to restriction fragments from the genomic termini predicted to contain covalently-linked protein; digestion of DNA with proteinase K restores band intensity. Full length gel (Fig. S3) is available in supplementary information. (B) Genome maps of phage Toil, enterobacterial phage PRD1 and Bacillus phage Bam35c. Heavy black lines represent the DNA molecules, with boxes above the lines representing genes transcribed from the forward strand and boxes below the lines representing genes transcribed from the reverse strand. Predicted functions of major genes are annotated: DNA polymerase (DNA pol), LysM domain-containing protein (lysM), lytic transglycosylase used for phage DNA entry (entry SLT), CHAP domain-containing protein (CHAP), receptor-binding protein (rbp), and predicted endolysin (lysA). ORF numbers or gene names are indicated for genes with annotated functions. Black arrows on the phage Toil map represent the positions of HindIII sites.