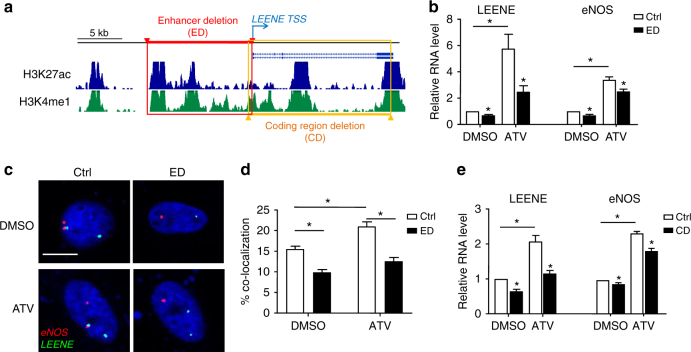
Fig. 4.
Gene editing of LEENE locus influences eNOS transcription. a Schematic illustration of CRISPR-Cas9 targeting strategy. Regions in red and orange indicate, respectively, the upstream enhancer/promoter region or the coding region deleted by sgRNA-guided Cas9, resulting in enhancer deletion (ED) and coding region deletion (CD) in LEENE locus. b LEENE and eNOS RNA levels in ECs transfected with control Cas9 plasmid (Ctrl) or “ED” Cas9-sgRNAs were quantified using qPCR. c DNA FISH for proximity association of LEENE and eNOS genomic loci. ECs transfected with control (Ctrl) or “ED” Cas9-sgRNAs were treated with DMSO or ATV (1 μM) for 24 h. Scale bar = 10 μm. d Percentage of cells with LEENE and eNOS proximity association (distance <1 μm). n = 678 in “DMSO-Ctrl” group; n = 425 in “DMSO-ED” group; n = 632 in “ATV-Ctrl” group; n = 581 in “ATV-ED” group. e qPCR quantification of LEENE and eNOS RNA levels in ECs transfected with control Cas9 (Ctrl) or “CD” Cas9-sgRNAs. All data are presented as mean ± SEM. n = 5 in each group unless specified. *p < 0.05 compared with “Ctrl” or between indicated groups based on Student’s t test