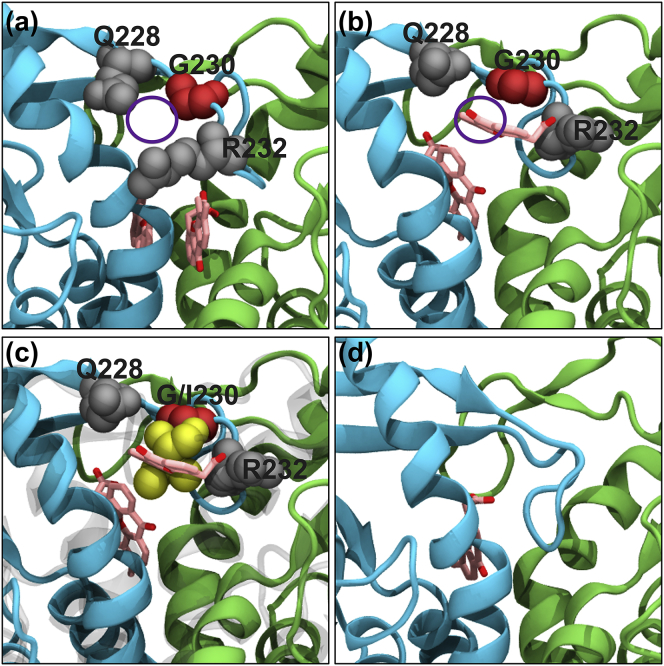
Figure 5.
Snapshots from a MD simulation of WT hSTING with DMXAA. (a) At the beginning of the simulation (0 ns), there is a gap in the lid region that results from converting the I230 residue in the PDB: 4QXP crystal structure into WT hSTING with a glycine (highlighted in red). (b) Twenty-eight nanoseconds into the simulation, one of the DMXAA molecules exits the binding site exactly at the opening created by the I230G mutation. (c) An overlay of the I230 residue from a separate MD simulation shows the extra bulk from the isoleucine residue would result in a steric barrier preventing DMXAA from exiting the binding site. (d) At the end of the 150-ns MD simulation, one of the DMXAAs has exited the binding site and the tip of the lid region has come down and filled the space previously occupied by the compound.