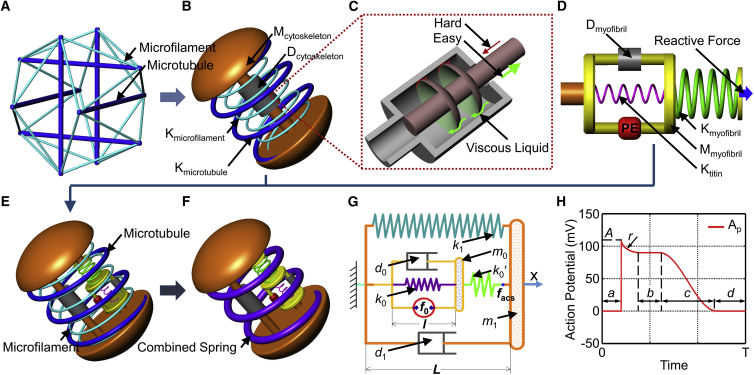
Figure 1.
Mechanical model of the cytoskeleton and the active contractile structure of a cardiomyocyte. (A) Tensegrity model, where the thick and thin lines represent the microtubules and microfilaments, respectively. (B) Mechanical model of cytoskeleton structure, where the Kmicrotubule represents the microtubules with a spring constant of kmicrotubule, and Kactin represents the microfilaments with a spring constant of kactin, Mcytoskeleton provides the inertial force induced by variable motion with a parameter of m1, and Dcytoskeleton provides the resistance against the deformation velocity of the cell with a damping coefficient of d1. (C) Schematic diagram of the damper with a variable damping coefficient. (D) Mechanical model of sarcomeres (myofibril) as the active contractile structure of a cardiomyocyte: Dmyofibril provides the resistance during sarcomere contraction with a damping coefficient of d0; Ktitin provides the static passive tension of the cell with a spring constant of k0; Mmyofibril describes the equivalent mass of the myofibrils with a parameter of m0; PE is the power element of the cardiomyocyte; Kmyofibril can transmit the contraction force from the sarcomere to the cytomembrane, and its spring constant is k0′; the reactive force is the resistance of the cytoskeleton. (E) Mechanical model of a cardiomyocyte. (F) Simplified mechanical model of a cardiomyocyte. (G) Two-dimensional mechanical model of a cardiomyocyte: k1 is the parameter of the integrated spring constant including the microtubule, microfilaments, and substrate-induced elasticity; l is the length of the sarcomere with an original value (relaxed phase) of l0; L is the length of the cell with an original value (relaxed phase) of L0; facs is the force of the active contractile structure of the cardiomyocyte. (H) Schematic model of internal relative action potential triggering the cell to regularly contract, where A, a, b, c, d, and r are the parameters to define the relative action potential pattern and will be identified based on the cellular beating patterns measured with SICM. To see this figure in color, go online.