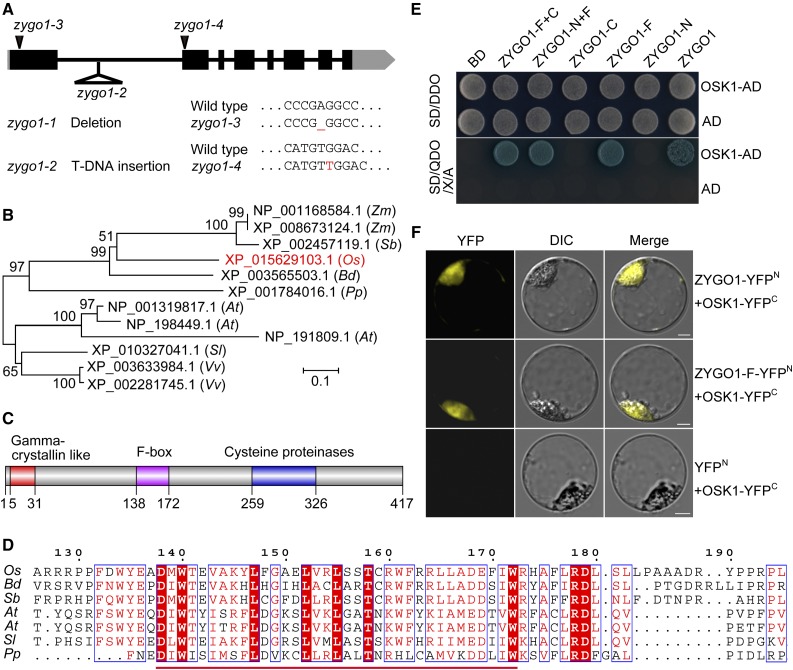
Figure 1.
ZYGO1 Encodes a Novel F-Box Protein in Rice.
(A) Gene structure of ZYGO1 and the mutation sites of zygo1 mutants. Black blocks and lines represent exons and introns, respectively. Untranslated regions are shown as gray boxes.
(B) An unrooted tree based on the full-length protein sequences of ZYGO1 and its homologs from seven plant species. ZYGO1 is shown in red. Zm, Zea mays; Sb, Sorghum bicolor; Os, Oryza sativa; Bd, Brachypodium distachyon; Pp, Physcomitrella patens; At, Arabidopsis thaliana; Sl, Solanum lycopersicum; Vv, Vitis vinifera.
(C) ZYGO1 contains a gamma-crystallin-like motif, an F-box domain, and a cysteine proteinase motif.
(D) Multiple sequence alignment of the F-box domain of ZYGO1 with its homologs from five plant species. Amino acid sequence of the F-box domain is underlined and conserved amino acids are highlighted in red.
(E) ZYGO1 interacts with OSK1 in yeast two-hybrid assays. Interactions were verified by the growth and the blue color of yeast cells on SD/QDO/X/A (SD-Ade-His-Leu-Trp + X-α-gal + Aureobasidin) medium. BD, bait vector; AD, prey vector; ZYGO1-N, the N-terminal residues (amino acids 1–137) of ZYGO1; ZYGO1-F, the F-box domain of ZYGO1 (amino acids 138–172); ZYGO1-C, the C-terminal residues (amino acids 173–417) of ZYGO1; ZYGO1-N+F, 1 to 172 amino acids of ZYGO1; ZYGO1-F+C, 138 to 417 amino acids of ZYGO1.
(F) BiFC assays showing the interaction between OSK1 and the full-length protein sequence or the F-box domain of ZYGO1 in rice protoplasts. Bars = 5 µm.