The Arabidopsis MOS4-associated complex promotes microRNA biogenesis and precursor messenger RNA splicing.
Abstract
In Arabidopsis thaliana, the MOS4-ASSOCIATED COMPLEX (MAC) is required for defense and development. The evolutionarily conserved, putative RNA helicase MAC7 is a component of the Arabidopsis MAC, and the human MAC7 homolog, Aquarius, is implicated in pre-mRNA splicing. Here, we show that mac7-1, a partial loss-of-function mutant in MAC7, and two other MAC subunit mutants, mac3a mac3b and prl1 prl2 (pleiotropic regulatory locus), exhibit reduced microRNA (miRNA) levels, indicating that MAC promotes miRNA biogenesis. The mac7-1 mutant shows reduced primary miRNA (pri-miRNA) levels without affecting miRNA gene (MIR) promoter activity or the half-life of pri-miRNA transcripts. As a nuclear protein, MAC7 is not concentrated in dicing bodies, but it affects the localization of HYPONASTIC LEAVES1 (HYL1), a key protein in pri-miRNA processing, to dicing bodies. Immunoprecipitation of HYL1 retrieved 11 known MAC subunits, including MAC7, indicating association between HYL1 and MAC. We propose that MAC7 links MIR transcription to pri-miRNA processing. RNA-seq analysis showed that downregulated genes in MAC subunit mutants are mostly involved in plant defense and stimulus responses, confirming a role of MAC in biotic and abiotic stress responses. We also discovered global intron retention defects in mutants in three subunits of MAC, thus linking MAC function to splicing in Arabidopsis.
INTRODUCTION
MicroRNAs (miRNAs) are a class of small RNAs that are ∼20 to 24 nucleotides in length and act as posttranscriptional regulators of gene expression in both animals and plants. miRNAs are processed from hairpin-containing precursors, primary miRNAs (pri-miRNAs), by RNase III family enzymes. A mature miRNA is loaded into an Argonaute protein to form a silencing complex and guides this silencing complex to target RNAs through sequence complementarity with target RNAs to result in their degradation or translational repression (Rogers and Chen, 2013).
The plant miRNA pathway has been intensively studied in the past decade. Early studies identified key proteins with catalytic activities, including RNA polymerase II (Pol II) that transcribes miRNA genes (MIR) (Xie et al., 2005; Zheng et al., 2009); DICER-LIKE1 (DCL1), an RNase III family enzyme excising miRNAs from stem-loop precursors; HUA ENHANCER1 (HEN1), a methyltransferase stabilizing miRNAs by 2’-O-methylation (Park et al., 2002; Li et al., 2005; Yu et al., 2005, 2010; Yang et al., 2006b); and ARGONAUTE1 (AGO1), the enzyme mediating miRNA-guided target RNA cleavage (Baumberger and Baulcombe, 2005; Ji et al., 2011; Carbonell et al., 2012; Arribas-Hernández et al., 2016). Many factors that assist in the transcription and processing steps of miRNA biogenesis have been identified (Rogers and Chen, 2013; Achkar et al., 2016). Pol II-mediated MIR transcription requires the transcriptional coactivator Mediator (Kim et al., 2011) and the transcription factor NEGATIVE ON TATA LESS2 (NOT2) (Wang et al., 2013) and is regulated by CYCLIN-DEPENDENT KINASEs (Hajheidari et al., 2012). In addition, the DNA binding protein CELL DIVISION CYCLE5 (CDC5) (Zhang et al., 2013b) and Elongator (Fang et al., 2015) also regulate MIR transcription by Pol II.
While interacting with Pol II, NOT2, CDC5, and Elongator also interact with DCL1 and several DCL1-interacting proteins and probably recruit them to pri-miRNAs to facilitate their processing (Wang et al., 2013; Zhang et al., 2013b; Fang et al., 2015). During the processing of miRNA precursors, DCL1 forms a dicing complex with the dsRNA binding domain protein HYPONASTIC LEAVES1 (HYL1/DRB1) and the zinc finger protein SERRATE (SE) (Han et al., 2004; Kurihara and Watanabe, 2004; Kurihara et al., 2006; Yang et al., 2006a; Dong et al., 2008). DCL1 and HYL1 are enriched in subnuclear bodies, referred to as dicing bodies, which are considered to be sites of miRNA precursor processing (Han et al., 2004; Fang and Spector, 2007; Song et al., 2007). Many other proteins also interact with DCL1 directly or indirectly in miRNA biogenesis, such as cap binding proteins (Laubinger et al., 2008; Raczynska et al., 2014), the forkhead-associated domain containing protein DAWDLE (Yu et al., 2008), the G-patch domain containing RNA binding protein TOUGH (Ren et al., 2012), and the WD-40 protein PLEIOTROPIC REGULATORY LOCUS1 (PRL1) (Zhang et al., 2014). In addition, several proteins act in miRNA biogenesis through the regulation of HYL1 phosphorylation, such as C-TERMINAL DOMAIN PHOSPHATASE-LIKE proteins and a K homology domain protein REGULATOR OF CBF GENE EXPRESSION3 (Manavella et al., 2012; Karlsson et al., 2015). Several other factors, including two core members of the THO/TREX complex, THO2 and EMU (Furumizu et al., 2010; Francisco-Mangilet et al., 2015), and the RNA binding protein MODIFIER OF SNC1,2 (MOS2) (Wu et al., 2013) do not seem to interact with any known dicing complex components, but still affect miRNA biogenesis.
Among the known miRNA biogenesis factors, CDC5 and PRL1 belong to the same complex, the MOS4-associated complex (MAC). MAC is a highly conserved complex among eukaryotes, with its orthologs known as the NINETEEN COMPLEX (NTC) or Prp19 complex (Prp19C) in yeast and humans. The Arabidopsis thaliana MAC, yeast, and human NTC/Prp19C all associate with the spliceosome and are predicted to share conserved functions in splicing in all three systems (Monaghan et al., 2009; Johnson et al., 2011; Koncz et al., 2012; Deng et al., 2016). Arabidopsis CDC5 is a MYB-related transcription factor and PRL1 is a conserved nuclear WD-40 protein. As core components of MAC, they regulate plant development and immunity through molecular mechanisms that remain unclear (Németh et al., 1998; Lin et al., 2007; Palma et al., 2007). They both promote miRNA biogenesis but may have distinct molecular functions. CDC5 binds to MIR promoters and interacts with DCL1 and SE to enhance miRNA biogenesis (Zhang et al., 2013b), while PRL1 may stabilize pri-miRNAs through its RNA binding activity and enhance DCL1 activity (Zhang et al., 2014). Other Arabidopsis MAC subunits include two homologous proteins MAC3A and MAC3B (MAC3B was shown to have E3 ligase activity in vitro), MAC7/Aquarius (an RNA helicase), and more than 10 other members (Wiborg et al., 2008; Monaghan et al., 2009, 2010; Koncz et al., 2012).
Arabidopsis MAC7 is a putative RNA helicase, which is highly conserved in eukaryotes. The human MAC7 homolog is the intron binding protein Aquarius (IBP160/AQR), which has ATPase and RNA helicase activities. It associates with the spliceosome and contributes to efficient precursor-mRNA splicing in vitro (Hirose et al., 2006; De et al., 2015). Recent publications show that EMB-4, the Caenorhabditis elegans homolog of MAC7, physically interacts with germline AGOs. It participates in the nuclear RNAi pathway and maintains the homeostasis of germline transcriptome in worms (Akay et al., 2017; Tyc et al., 2017). MAC7 was predicted to be an essential gene for embryo development (EMB gene) in Arabidopsis based on sequence similarity with EMB genes found in other eukaryotes (Tzafrir et al., 2004). However, the molecular function of MAC7 remains unknown.
In this study, we performed a genetic screen using a miRNA activity reporter line, the pSUC2:amiR-SUL (amiR-SUL) transgenic line (de Felippes et al., 2011). The expression of SUCROSE-PROTON SYMPORTER2 (SUC2) promoter-driven artificial miRNA targeting the CHLORINA42 (CH42) gene creates a bleached phenotype along the leaf veins. This bleaching of mesophyll cells caused by the silencing effects of the artificial miRNA results in an easily scorable phenotype reflecting miRNA activities in plants (de Felippes et al., 2011). From this screen, we identified a point mutation in MAC7, mac7-1, as a miRNA activity suppressor mutant. We showed that MAC7 affects miRNA accumulation through promotion of pri-miRNA biogenesis in Arabidopsis. We found that in other MAC subunit mutants, including mac3a mac3b and prl1 prl2 double mutants, miRNA biogenesis is also compromised, indicating that MAC participates in miRNA biogenesis as a complex. Consistent with this, HYL1 immunoprecipitation mass spectrometry analyses revealed that HYL1 associates with MAC in vivo. In addition, we uncovered global intron retention defects in mac7-1, mac3a mac3b, and prl1 prl2 mutants through RNA-seq analysis. Our molecular characterization of MAC7 and its associated MAC components revealed their functions in miRNA biogenesis and pre-mRNA splicing, which could possibly explain their roles in plant development and stress responses.
RESULTS
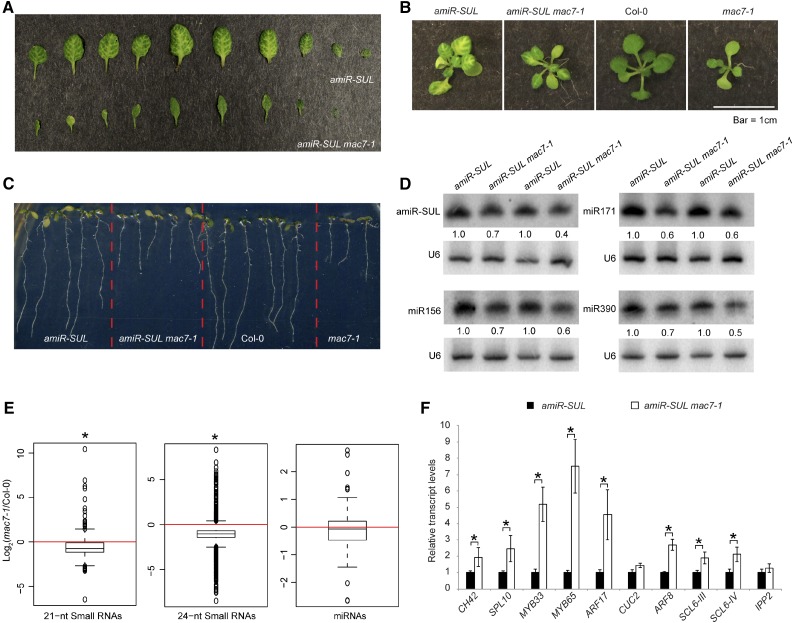
A Silencing Suppressor Mutant Exhibits Pleiotropic Phenotypes and Reduced miRNA Levels
To identify new players of the miRNA pathway, we performed an ethyl methanesulfonate (EMS) mutagenesis screen using the amiR-SUL line (de Felippes et al., 2011). From this screen, we identified a mutant, which we named mac7-1 based on subsequent characterization (amiR-SUL mac7-1) with a reduced area of bleaching along the veins, indicating compromised miRNA activity (Figure 1A). The mutant has pleiotropic phenotypes, such as pointed leaves, reduced root length, reduced number of lateral roots, smaller plant stature, and reduced fertility (Figures 1B and 1C; Supplemental Figure 1). We crossed amiR-SUL mac7-1 with wild-type (Col-0) plants to remove the amiR-SUL transgene. We found that the mac7-1 mutant shows the same range of phenotypes in the Col-0 background as in the amiR-SUL background (Figures 1B and 1C; Supplemental Figure 1).
Figure 1.
A Silencing Suppressor Mutant mac7-1 Exhibits Pleiotropic Phenotypes and Compromised miRNA Accumulation.
(A) Differences in vein-centered bleaching in 3- to 4-week-old pSUC2:amiR-SUL (amiR-SUL) and amiR-SUL mac7-1 seedlings.
(B) and (C) Morphological phenotypes of amiR-SUL, amiR-SUL mac7-1, Col-0, and mac7-1 plants. Images of rosettes and roots were taken from 2- to 3-week-old and 1-week-old plants, respectively.
(D) RNA gel blotting analysis of miRNAs from amiR-SUL and amiR-SUL mac7-1 inflorescences. The miRNA signals were quantified and normalized to those of U6, and values were relative to amiR-SUL (set to 1). Two biological replicates of inflorescences collected from plants grown separately but under the same conditions were processed and shown.
(E) Global abundance of 21- and 24-nucleotide small RNAs and miRNAs in Col-0 and mac7-1 as determined by small RNA-seq. Small RNA libraries were generated from 12-d-old seedlings growing on MS plates. The normalization of small RNAs was against 45S rRNA reads and abundance was expressed as RPMR (reads per million of 45S rRNA reads), and log2 ratios of mac7-1/Col-0 were plotted. Asterisks indicate that the mean is significantly below 0 (Wilcoxon test, P < 2.2e-16).
(F) Determination of miRNA target mRNA levels in amiR-SUL and amiR-SUL mac7-1 using 12-d-old seedlings by RT-qPCR. The housekeeping gene IPP2 was included as a control. Expression levels were normalized to those of UBIQUITIN5 (UBQ5) and compared with those in amiR-SUL (set to 1). Error bars indicate sd from three technical replicates. Asterisks indicate significant difference between Col-0 and mac7-1 (t test, P < 0.05).
Because the mutant shows compromised amiR-SUL activities, we speculated that reduced miRNA accumulation could be a reason. We performed RNA gel blot analyses to detect amiR-SUL as well as many endogenous miRNAs. We found that amiR-SUL, miR156, miR171, miR390, and many other miRNAs showed reduced accumulation in both inflorescences and seedlings. The reduction in miRNA abundance was small but reproducible in several biological replicates (Figure 1D; Supplemental Figure 2). To assess the global influence of the mutation on small RNAs, we also performed small RNA-seq with Col-0 and mac7-1 seedlings. We found that 21- and 24-nucleotide small RNAs, which represent the two most abundant small RNA size classes, showed a significant global reduction in the mac7-1 mutant. miRNAs showed a slight global reduction in the mac7-1 mutant but the reduction was not statistically significant (Figure 1E; Supplemental Data Set 1). Most miRNAs that were found to show reduced abundance by RNA gel blotting also had lower levels in mac7-1 in small RNA-seq, although a few miRNAs did not (e.g., miR156 and miR164) (Supplemental Figure 2C). The minor inconsistency between RNA gel blotting and small RNA-seq could be caused by technical limitations in small RNA-seq, e.g., bias in RNA adaptor ligation or PCR amplification. Since small RNA-seq entails more procedures that are prone to bias, we believe that the RNA gel blotting results are more accurate.
We examined the expression of CH42 and eight known miRNA target genes in amiR-SUL and amiR-SUL mac7-1. Opposite to the reduced miRNA accumulation, the expression levels of CH42 and seven endogenous miRNA targets were upregulated in amiR-SUL mac7-1 (Figure 1F).
The reduced bleaching phenotype in amiR-SUL mac7-1 might be attributed to other factors other than, or in addition to, reduced amiR-SUL activity, e.g., enhanced transcription of CH42. We performed RT-qPCR and immunoblotting to examine CH42 mRNA and protein levels, respectively, in Col-0 and mac7-1 to determine whether an amiR-SUL-independent effect of the mac7-1 mutation was present. While CH42 mRNA and protein levels were both increased in amiR-SUL mac7-1 in comparison to amiR-SUL, no difference was found between Col-0 and mac7-1 (Supplemental Figure 3), indicating that the reduced bleaching phenotype and elevated CH42 expression in amiR-SUL mac7-1 was most likely caused by changes in amiR-SUL activity only.
A Point Mutation in MAC7 Is Responsible for the Suppression of amiR-SUL-Induced Silencing
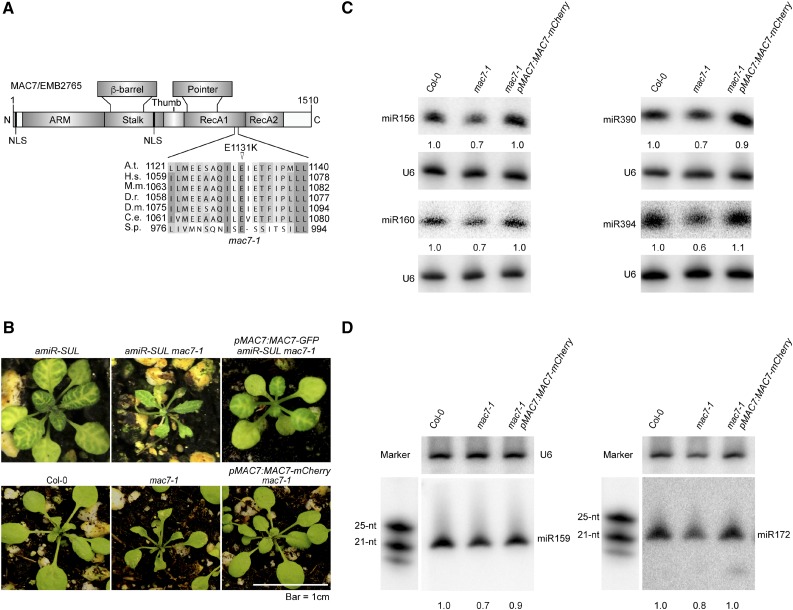
Through genome resequencing of pooled F2 mutants from a backcross between amiR-SUL mac7-1 and the parental amiR-SUL line, we found a C-to-T nucleotide transition that changes a conserved glutamic acid to lysine (E1131K) in the open reading frame of At2g38770 (MAC7/EMB2765) (Figure 2A). To determine whether this mutation was the causal mutation that suppressed amiR-SUL, we first examined the linkage between the mutation and the visible phenotype. In the F2 population of the backcross, we identified 84 plants with the mac7-1 phenotype, i.e., reduced vein-centered leaf bleaching. These plants were genotyped for the C-to-T mutation in MAC7. All 84 plants were found to be homozygous for this mutation; thus, the amiR-SUL suppressor phenotype was linked with this mutation.
Figure 2.
A Point Mutation in MAC7 Is Responsible for the Morphological and Molecular Phenotypes in the mac7-1 Mutant.
(A) A diagram of the MAC7 protein showing various domains, the predicted nuclear localization signal (NLS), and the E1131K mutation in the mac7-1 mutant. A sequence alignment of MAC7 and its orthologs in the region containing the E1131K mutation in the mac7-1 mutant is also shown. Abbreviations for species are as follows: Arabidopsis thaliana (A.t.), Homo sapiens (H.s.), Mus musculus (M.m.), Danio rerio (D.r.), Drosophila melanogaster (D.m.), Caenorhabditis elegans (C.e.), and Schizosaccharomyces pombe (S.p.). N, N terminus; ARM, armadillo domain; RecA, RecA-like domains; C, C terminus. The point mutation site is labeled by a triangle.
(B) Morphological phenotypes of 3- to 4-week-old seedlings of the indicated genotypes. pMAC7:MAC7-GFP and pMAC7:MAC7-mCherry were transformed into amiR-SUL mac7-1 and mac7-1, respectively.
(C) and (D) RNA gel blotting analysis of miRNAs from Col-0, mac7-1, and the complementation line mac7-1 pMAC7:MAC7-mCherry using inflorescences (C) and 12-d-old seedlings (D). The miRNA signals were quantified as described in Figure 1D. RNA markers (NEB, N2102S) shown in (D) were resolved in the same gel as miRNAs and probed separately by a DNA probe complementary to the marker sequences.
Next, to confirm that the causal mutation of the suppressor was in MAC7, we generated MAC7 promoter driven MAC7-GFP and MAC7-mCherry fusion constructs and introduced them into mac7-1 in amiR-SUL and Col-0 backgrounds. pMAC7:MAC7-GFP or pMAC7:MAC7-mCherry fully complemented the morphological defects (Figure 2B) and pMAC7:MAC7-mCherry restored the accumulation of miRNAs of the mutant in both inflorescences and seedlings (Figures 2C and 2D). Thus, the causal mutation in the suppressor was in MAC7, and we named this mutation mac7-1.
To obtain another mac7 allele, we ordered a T-DNA insertion line, SALK_129044 (which we named mac7-2) (Supplemental Figure 4A). Genotyping mac7-2 in the progeny of a selfed mac7-2/+ plant revealed 2:1 segregation between heterozygous and wild-type plants (Supplemental Table 1), suggesting that mac7-2 homozygous plants were embryo lethal. Unlike wild-type siliques that contained only normal-looking green seeds, siliques from heterozygous (mac7-2/+) plants had aborted seeds that appeared pale white (Supplemental Figure 4B). An approximate 3:1 ratio between normal seeds and aborted seeds was found (Supplemental Table 1), which was a strong indication of embryo lethality of the homozygous mutant. In addition, the viability of pollen from mac7-2/+ plants appeared normal (Supplemental Figure 4C). The above evidence demonstrated that mac7-2 is a recessive, embryo-lethal mutation. As mac7-1 homozygous plants are viable, this mutation is likely a partial loss-of-function allele.
The MAC7 gene encodes an RNA helicase conserved from yeast to animals and plants. The structure of the human MAC7 ortholog, Aquarius, has been determined (De et al., 2015). Based on homology modeling, we predict that the plant MAC7 also has a long N-terminal domain composed of armadillo repeats (ARM), which is crucial for protein-protein interactions, a stalk and a β-barrel domain, which has an architectural role, a thumb and a pointer domain, which may participate in interactions with other proteins, and two RecA-like domains (RecA1 and RecA2), which form a motor module required for ATP hydrolysis, RNA unwinding, and the coupling of these two processes. The mac7-1 mutation leads to a change of a highly conserved amino acid in the RecA1 domain, which very likely affects the core functions of this protein in ATP binding/hydrolysis and nucleic acid unwinding (Figure 2A; Supplemental Figure 5) (De et al., 2015; Ozgur et al., 2015).
MAC7 Promotes pri-miRNA Levels without Affecting MIR Promoter Activities or pri-miRNA Transcript Half-Life
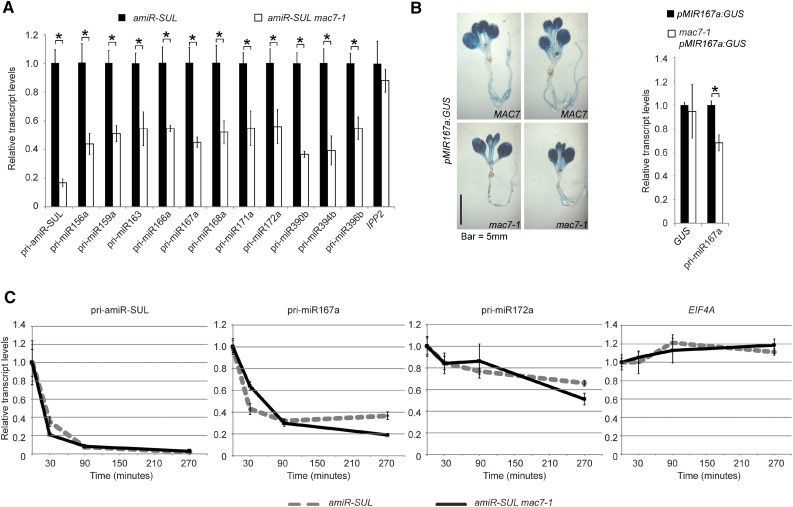
To determine the molecular mechanisms of MAC7 in miRNA biogenesis, we examined the levels of pri-miRNAs. We found that the levels of pri-miRNAs from many MIR genes were reduced in the mac7-1 mutant (Figure 3A). This reduction could be attributed to reduced transcription of MIR genes or enhanced pri-miRNA degradation or processing. We first examined whether mac7-1 affects MIR promoter activities. We crossed mac7-1 to a miRNA promoter reporter line, pMIR167a:GUS, in which the transgene was inserted into a single genomic locus. We then examined GUS activity by staining and GUS transcripts level by real-time RT-PCR in wild-type and mac7-1 plants, in which the transgene was homozygous. There was no detectable difference between the wild type and mac7-1 in terms of GUS activity or GUS transcripts level, indicating that mac7-1 did not affect miRNA promoter activities (Figure 3B). We also examined whether the reduction of amiR-SUL level in mac7-1 was due to reduced SUC2 promoter activity. If this was the case, we would expect the endogenous SUC2 RNA to be reduced in abundance in mac7-1. Real-time RT-PCR revealed that SUC2 mRNA levels did not change significantly in mac7-1, implying that SUC2 promoter activity was not affected by mac7-1 and the reduction in amiR-SUL levels was not due to impaired SUC2 promoter activities in the mutant (Supplemental Figure 6A).
Figure 3.
MAC7 Promotes pri-miRNA Production.
(A) Determination of pri-miRNA levels in amiR-SUL and amiR-SUL mac7-1 inflorescences by RT-qPCR. The housekeeping gene IPP2 was included as a control. Expression levels were normalized to those of UBQ5 and compared with those in amiR-SUL (set to 1). Error bars indicate sd from three technical replicates. Asterisks indicate significant difference between amiR-SUL and amiR-SUL mac7-1 (t test, P < 0.05).
(B) Representative GUS staining images of pMIR167a:GUS and mac7-1 pMIR167a:GUS seedlings. The transcript levels of GUS and endogenous pri-miR167a in pMIR167a:GUS and mac7-1 pMIR167a:GUS seedlings were determined by RT-qPCR. Expression levels were normalized to those of UBQ5 and compared with those in pMIR167a:GUS (set to 1). Error bars indicate sd from three technical replicates. Asterisks, t test P < 0.05.
(C) Half-life measurements for pri-amiR-SUL, pri-miR167a, pri-miR172a, and EIF4A mRNA. Two-week-old amiR-SUL and amiR-SUL mac7-1 seedlings were treated with 0.6 mM cordycepin and harvested at various time points. RT-qPCR was performed to determine the levels of various pri-miRNAs and EIF4A mRNA. UBQ5 served as an internal control. Values at time 0 were set to 1. Error bars indicate sd from three technical replicates. Two biological replicates were performed and similar results were obtained.
Next, we measured the half-lives of pri-miRNAs in amiR-SUL and mac7-1 amiR-SUL. Seedlings were treated with the transcription inhibitor cordycepin, and RNAs were isolated at different time points. The levels of pri-amiR-SUL, pri-miR167a, and pri-miR172a were determined by real-time RT-PCR. Similar half-lives were found for these pri-miRNAs in the two genotypes (Figure 3C).
It is also possible that MAC7 affects miRNA biogenesis indirectly through affecting the expression of key miRNA biogenesis factors. To test this possibility, we determined transcript and protein levels of several key miRNA biogenesis factors (i.e., DCL1, HYL1, SE, AGO1, HEN1 [transcript only]) in amiR-SUL and amiR-SUL mac7-1. No differences were detected for any of the genes in the two genotypes (Supplemental Figures 6B to 6 D).
MAC7 Is a Nuclear Protein Interacting with Other MAC Components, Which Are Also Required for miRNA Biogenesis
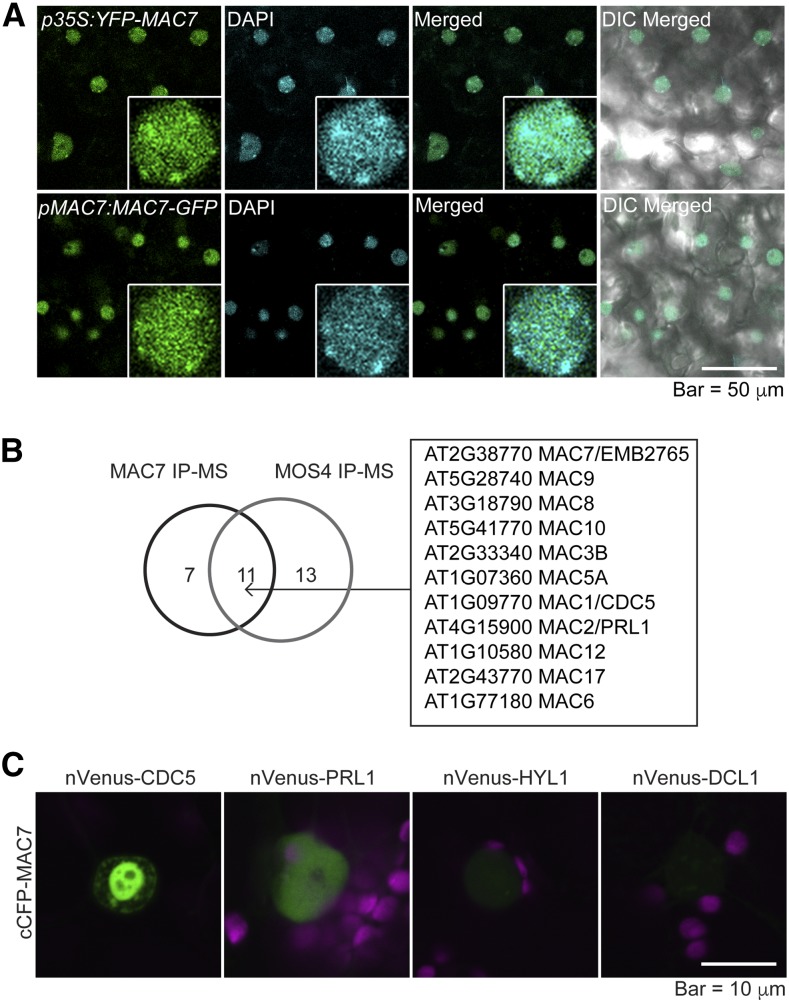
To further characterize the molecular functions of the MAC7 protein, we examined the subcellular localization of fluorescent protein tagged MAC7. Transgenic plants expressing p35S:YFP-MAC7 or pMAC7:MAC7-GFP exhibited nuclear GFP signals (Figure 4A), which is consistent with the presence of a predicted nuclear localization signal in MAC7 (Figure 2A). To determine if MAC7 localized in dicing bodies, we transiently expressed YFP-MAC7 and DCL1-YFP in tobacco (Nicotiana benthamiana) leaves and compared their expression patterns. While DCL1-YFP showed weak nucleoplasmic signals and strong dicing body signals, YFP-MAC7 exhibited dispersed distribution in the nucleoplasm and was absent from the nucleolus (Supplemental Figure 7A). When YFP-MAC7 was coexpressed with TagRFP-HYL1, signals from the two proteins overlapped in the nucleoplasm while only TagRFP-HYL1 was found concentrated in dicing bodies (Supplemental Figure 7B). Immunoblot analyses confirmed the expression of GFP- or YFP-tagged MAC7 in Arabidopsis transgenic lines (Supplemental Figure 7C) and in infiltrated tobacco leaves (Supplemental Figure 7D). These data suggest that MAC7 does not display the dicing body patterns as DCL1 and HYL1 do. The dispersed nucleoplasmic distribution of MAC7 suggests broader roles than miRNA biogenesis.
Figure 4.
MAC7 Is a Nuclear Protein Associated with Other MAC Subunits.
(A) Subcellular localization of N-terminal (p35S:YFP-MAC7) or C-terminal (pMAC7:MAC7-GFP) fluorescent protein tagged MAC7 in young leaves of Arabidopsis transgenic lines. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) and pseudo-colored in cyan. Enlarged nuclei are shown in the insets.
(B) MAC subunits identified from both MAC7 and MOS4 immunoprecipitation followed by mass spectrometry analysis (IP-MS).
(C) BiFC analysis of MAC7 with CDC5, PRL1, HYL1, and DCL1. Paired cCFP- and nVenus-fusion proteins were coinfiltrated into tobacco leaves. The BiFC signal (YFP) was detected at 48 h after infiltration by confocal microscopy, and was pseudo-colored in green. Magenta indicates autofluorescence of chlorophyll.
To uncover interacting partners of MAC7 and to determine whether MAC7 interacts with any known miRNA pathway proteins, we immunoprecipitated (IP) MAC7 and performed mass spectrometry (MS) analyses. In one experiment, IP was performed with Col-0 plants using anti-MAC7 antibodies with preimmune IgG as a negative control. In another experiment, IP was performed using Chromotek-RFP-Trap with a pMAC7:MAC7-mCherry mac7-1 line in which the transgene rescued the mutant phenotypes; IP was performed using the same RFP-Trap with Col-0 as a negative control. The two independent experiments consistently pulled down all major MAC components, demonstrating that MAC7 was part of the MAC (Supplemental Table 2). An overlapping set of proteins was identified from our MAC7 IP-MS and from published MOS4 IP-MS (Figure 4B; Supplemental Table 2) (Monaghan et al., 2009). Besides the known MAC subunits (Monaghan et al., 2009), seven additional proteins were discovered as potential MAC7-associated proteins (Supplemental Table 2), but the association between these proteins and MAC7, or whether they also belong to MAC, needs to be further investigated.
Previous studies showed that the MAC subunits CDC5 and PRL1 promote miRNA biogenesis (Zhang et al., 2013b, 2014). To further confirm the interactions between MAC7 with these two MAC subunits that are also miRNA biogenesis factors, we performed bimolecular fluorescence complementation (BiFC) analysis and found that MAC7 interacted with CDC5 and also interacted, albeit weakly, with PRL1 (Figure 4C). Similar BiFC studies did not reveal interactions between MAC7 and HYL1 or DCL1 (Figure 4C).
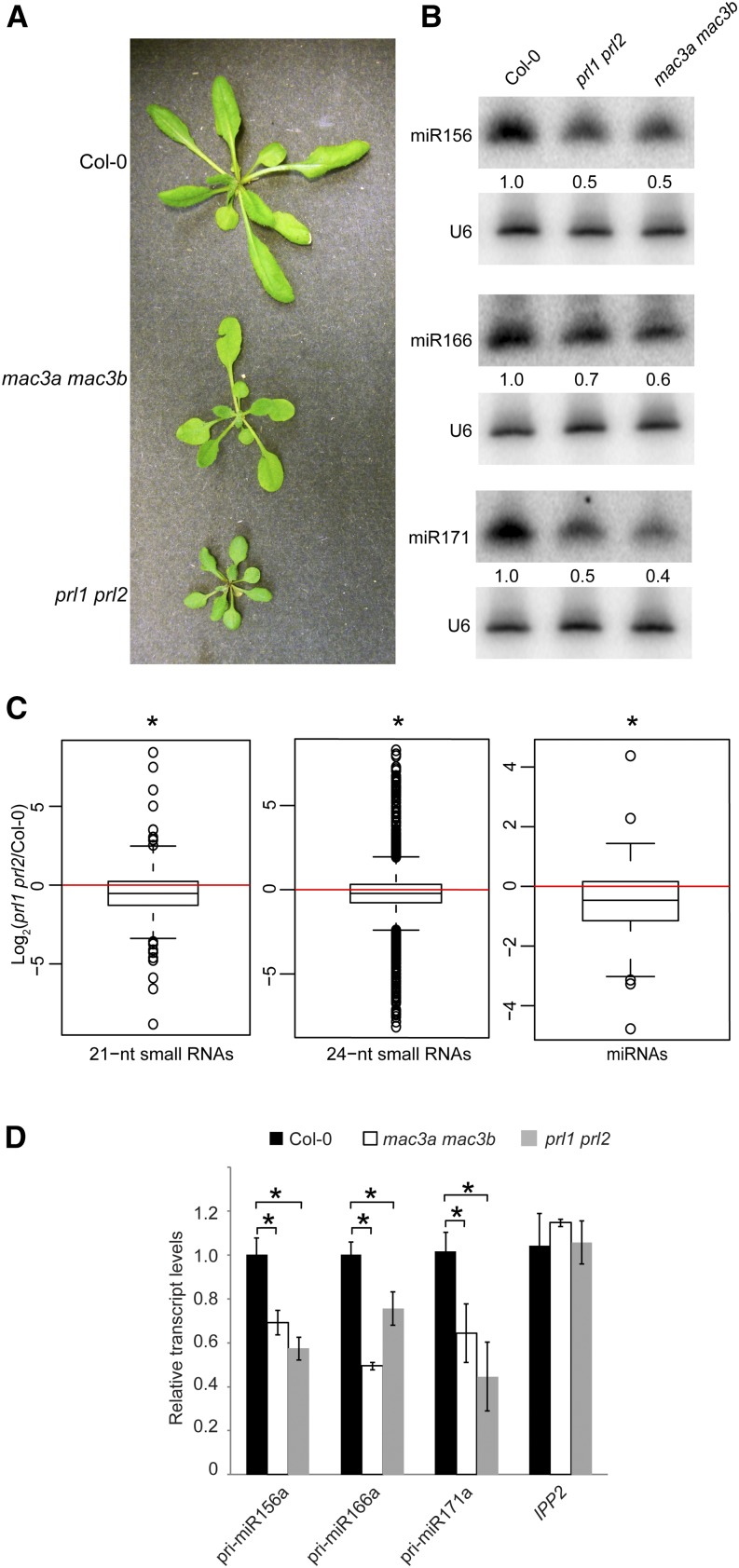
These results and our work on MAC7 raise the possibility that MAC plays a role in miRNA biogenesis, acting as a complex. To test this hypothesis, we examined whether core subunits of MAC are required for miRNA biogenesis. Arabidopsis MAC3B is a U-box E3 ubiquitin ligase and is a core MAC component (Monaghan et al., 2009). MAC3B has a homolog, MAC3A, which shares 82% identity with MAC3B at the amino acid level (Monaghan et al., 2009). PRL1 is also a core member of MAC. It has a homolog, PRL2, which is expressed at a much lower level than PRL1 (Weihmann et al., 2012). Previous studies showed that a prl1 single mutant has reduced miRNA levels but a mac3b mutant (SALK_050811) does not (Zhang et al., 2014). Several siRNAs, ta-siRNA255, and miRNA171 were found to show reduced accumulation in the mac3a mac3b (SALK_089300 and SALK_050811) double mutant (Zhang et al., 2013a). Here, we examined the morphological and molecular phenotypes of two double mutants, mac3a mac3b and prl1 prl2. Similar to other MAC subunit mutants, mac3a mac3b and prl1 prl2 exhibited pleiotropic developmental phenotypes (Figure 5A). RNA gel blot analyses showed that miR156, miR166, and miR171 all exhibited reduced accumulation in these double mutants (Figure 5B). We also performed small RNA-seq with Col-0 and prl1 prl2 seedlings to assess the global changes of small RNAs in prl1 prl2. Similar to those of mac7-1, 21- and 24-nucleotide small RNAs showed a global reduction in prl1 prl2. Unlike in mac7-1, a significant global reduction in miRNA levels was also found in prl1 prl2 (Figure 5C; Supplemental Data Set 1). This is consistent with mac7-1 being a weak allele representing a partial compromise in MAC function. RT-qPCR analyses revealed that the levels of pri-miRNAs were also reduced in mac3a mac3b and prl1 prl2 (Figure 5D). Thus, the molecular phenotypes of mac3a mac3b and prl1 prl2 mutants were similar to those of mac7-1.
Figure 5.
The MAC Subunit Genes MAC3a, MAC3b, PRL1, and PRL2 Also Promote miRNA Biogenesis.
(A) Morphological phenotypes of 4- to 5-week old plants of the indicated genotypes.
(B) miRNA levels in the wild type (Col-0) and indicated mutants as determined by RNA gel blotting. The miRNA signals were quantified and normalized to those of U6, and values were relative to Col-0 (arbitrarily set to 1). The RNA used in RNA gel blotting was extracted from the aerial parts of 12-d-old seedlings growing on MS agar plates.
(C) Global abundance of 21- and 24-nucleotide small RNAs and miRNAs in Col-0 and prl1 prl2 as determined by small RNA-seq. Small RNA libraries were generated from 12-d-old seedlings growing on MS plates. The normalization of small RNAs was against 45S rRNA reads and abundance was expressed as RPMR (reads per million of 45S rRNA reads), and log2 ratios of prl1 prl2/Col-0 were plotted. Asterisks indicate that the mean is significantly below 0 (Wilcoxon test, P < 2.2e-16).
(D) pri-miRNA levels in plants of the indicated genotypes as determined by RT-qPCR. The housekeeping gene IPP2 was included as a control. UBQ5 was used as an internal control and values in Col-0 were set to 1. Error bars indicate sd from three technical replicates, and asterisks indicate significant difference between Col-0 and the mutants (t test, P < 0.05).
MAC Interacts with HYL1
As CDC5 and PRL1 interact with dicing complex components (Zhang et al., 2013b, 2014), it is possible that MAC7, or the entire MAC, associates with the dicing complex. To explore the interactions between the dicing complex and MAC, we performed HYL1 IP-MS. In one experiment, IP was performed with a p35S:HYL1-YFP line and the negative control p35S:YFP transgenic line and Col-0 using Chromotek-GFP-Trap. In another experiment, IP was performed with Col-0 and the negative control hyl1-2 using anti-HYL1 antibodies. Eleven MAC subunits, including MAC7, were found in both IP-MS experiments (Supplemental Table 3), indicating that HYL1 associates with MAC in vivo.
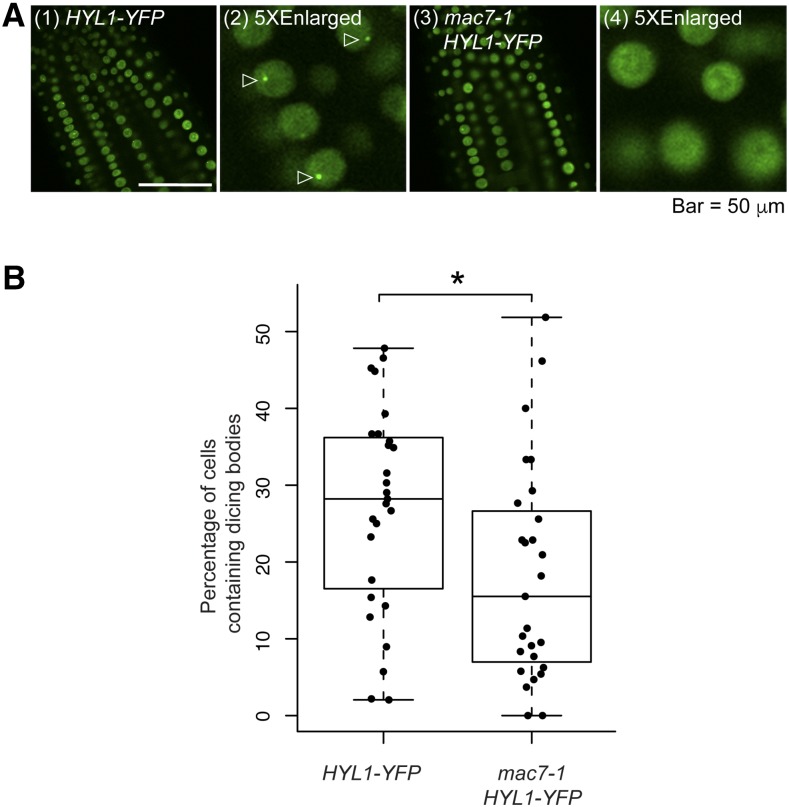
The interactions between HYL1 and MAC raised the possibility that MAC7 could be involved in pri-miRNA processing through affecting the dicing body localization of HYL1. We crossed mac7-1 to a HYL1-YFP transgenic line and quantified dicing body numbers in the wild type and mac7-1. The number of HYL1-YFP-positive dicing bodies was significantly reduced in mac7-1 compared with wild-type plants (Figures 6A and 6B), demonstrating that MAC7 is required for the proper localization of HYL1 in dicing bodies, which might partially explain the compromised miRNA levels in the mac7-1 mutant.
Figure 6.
MAC7 Affects HYL1 Subcellular Localization.
(A) Images of nuclei in root cells of 7- to 10-d-old seedlings of the indicated genotypes. Images (2) and (4) show five times enlarged images cropped from (1) and (3), respectively. Dicing bodies are indicated by triangles.
(B) The percentage of cells containing HYL1-positive dicing bodies in wild type and mac7-1. The quantification was performed by observing more than 1000 cells from 27 roots for each genotype. The asterisk indicates significant difference between the samples (t test, P < 0.05).
Downregulated Genes in mac Mutants Are Significantly Related to Stress Responses
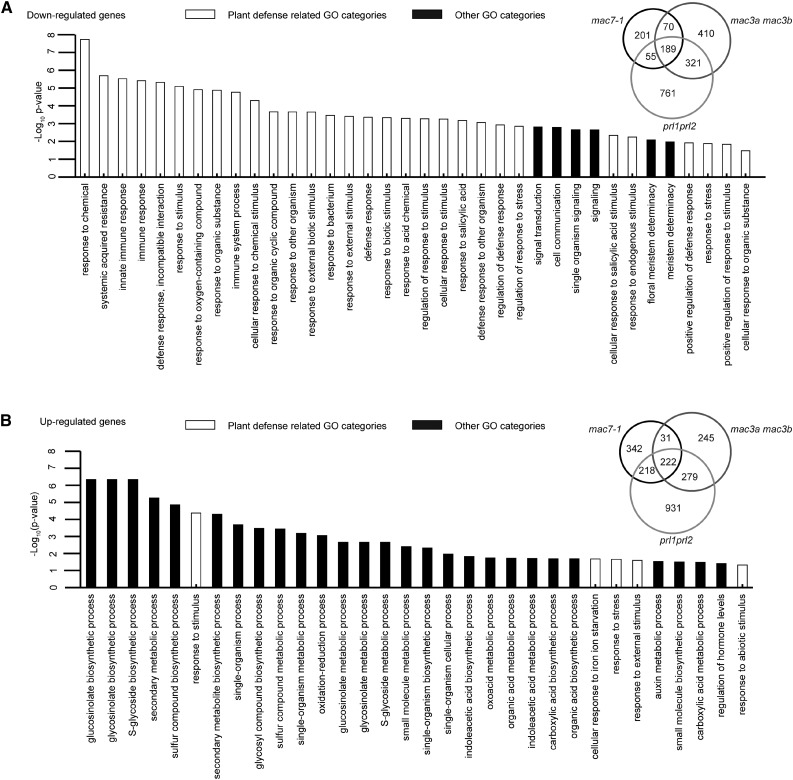
We also explored whether MAC plays a role in RNA metabolism in general. We performed RNA-seq with Col-0, mac7-1, mac3a mac3b, and prl1 prl2 seedlings in two biological replicates. Differentially expressed genes (DEGs) were identified between each mutant and wild type with fold change of 1.5 or more. Among the three mutants, we identified 2007 and 2268 downregulated (hypo-DEGs) and upregulated (hyper-DEGs) genes, respectively. A total of 189 hypo-DEGs and 222 hyper-DEGs were commonly found among mac7-1, mac3a mac3b, and prl1 prl2 (Figure 7; Supplemental Data Set 2). The overlap of the DEGs among these three mutants was significant (SuperExactTest, P = 0), indicating that these MAC components function as a complex and regulate the same group of genes. The large portion of nonoverlapped DEGs indicates that each subunit may also have its own function independent of MAC.
Figure 7.
Downregulated Genes in the mac Mutants Are Significantly Related to Stress Responses, and Upregulated Genes Are Involved in Various Biosynthetic and Metabolic Processes.
(A) GO enrichment analysis of 189 commonly downregulated genes in mac7-1, mac3a mac3b, and prl1 prl2. The degree of overlap among downregulated genes in mac7-1, mac3a mac3b, and prl1 prl2 is shown in the Venn diagram.
(B) GO enrichment analysis of 222 commonly upregulated genes mac7-1, mac3a mac3b, and prl1 prl2. The degree of overlap among upregulated genes in mac7-1, mac3a mac3b, and prl1 prl2 is shown in the Venn diagram.
To understand the biological functions of MAC, we examined the Gene Ontology (GO) terms enriched in the common DEGs (Gene Ontology Consortium, 2015). An enrichment of genes involved in stress responses, including stimulus responses, plant defense, or immune responses, was found in the hypo-DEGs. As for the hyper-DEGs, GO terms in various small molecule biosynthetic and metabolic processes were enriched (Figure 7; Supplemental Data Set 3). Many more GO terms related to stress responses were identified from the hypo-DEGs than the hyper-DEGs, implying that MAC tends to activate the expression of stress response genes. The results are consistent with previous findings showing that mac3a mac3b and prl1 prl2 double mutants are more susceptible to pathogen infection (Monaghan et al., 2009; Weihmann et al., 2012). To test whether MAC7 is also required for plant immunity, mac7-1 was infected with Pseudomonas syringae pv maculicola (P.s.m.) strain ES4326 together with Col-0 and prl1-2. The prl1 mutants are more susceptible to pathogen infection and therefore served as a positive control (Weihmann et al., 2012). Pathogen growth was assayed 3 d after bacterial inoculation. While P.s.m. ES4326 accumulated to higher levels in prl1-2 than in the wild type, mac7-1 was similar to the wild type in terms of bacterial titer (Supplemental Figure 8A). Perhaps this was due to mac7-1 being a weak allele, in which the magnitude of downregulation of the hypo-DEGs was small compared with that in other mac mutants (Supplemental Figure 8B).
MAC Subunits Affect pre-mRNA Splicing
Homologs of MAC in yeast and mammals play a critical role in pre-mRNA splicing. In Arabidopsis, we lack evidence for a widespread role of MAC in splicing; only the splicing patterns of several genes, such as SUPPRESSOR OF NPR1-1 CONSTITUTIVE1 and RESISTANCE TO PSEUDOMONAS SYRINGAE4, were shown to be altered in several MAC subunit mutants (Xu et al., 2012; Zhang et al., 2013a).
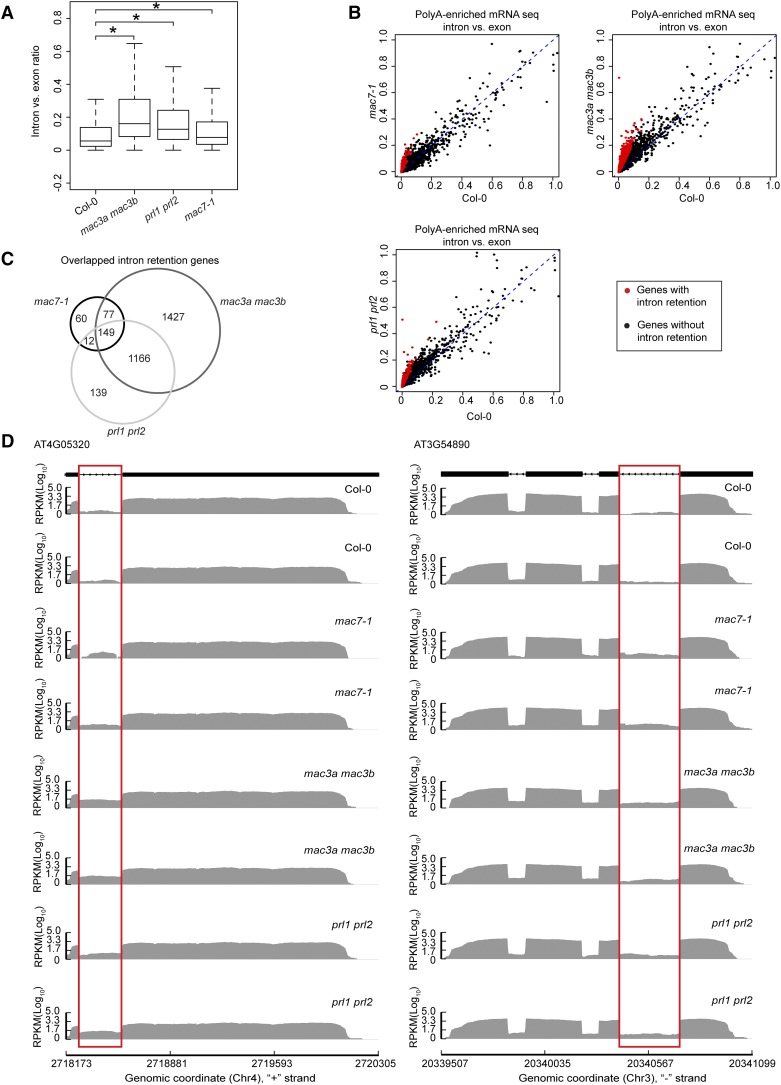
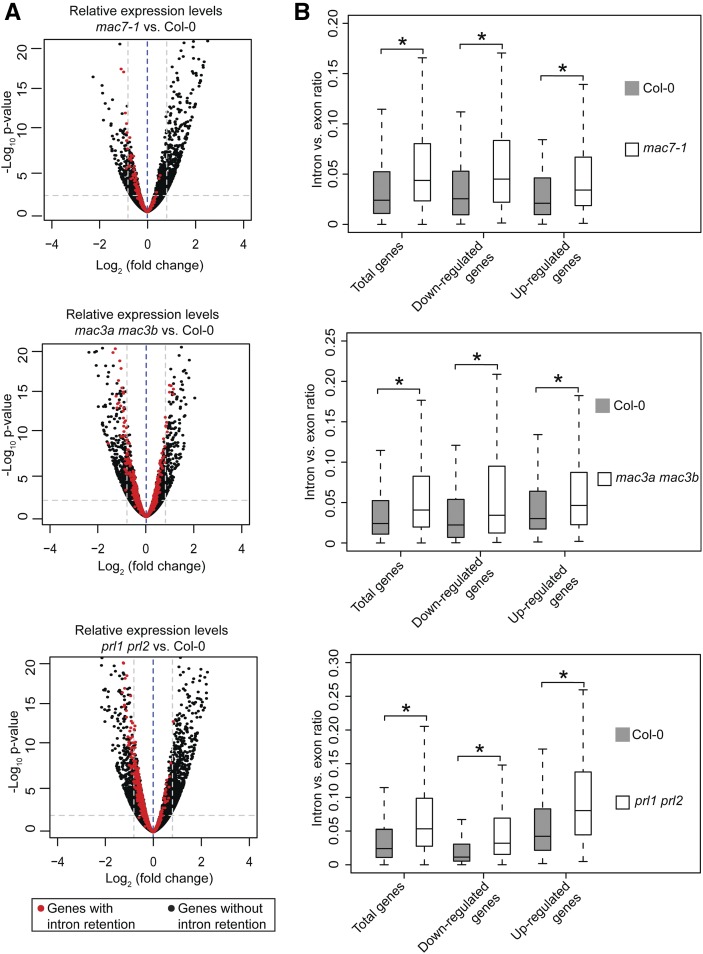
Since intron retention is a major form of alternative splicing in Arabidopsis (Ner-Gaon et al., 2004), we examined whether mac7-1, mac3a mac3b, and prl1 prl2 mutants had global intron retention defects. The ratio of RNA-seq reads mapping to introns (including 5′/3′ splice sites) and those mapping to exons only were used as a measure of intron retention (see Methods for details). All annotated transcripts were considered in sum as long as the read counts passed an abundance filter. All three mutants exhibited significantly higher levels of intron retention compared with Col-0 (Wilcoxon test, P < 2.2e-16) (Figure 8A; Supplemental Data Set 4). Next, using the ratio of intron reads versus exon reads as a measure of intron retention levels, we identified genes with intron retention defects in each mutant as compared with the wild type (see Methods for details). A total of 2819, 1466, and 298 genes were found to have intron retention defects in mac3a mac3b, prl1 prl2, and mac7-1, respectively (Figures 8B and 8C; Supplemental Data Set 4). Significant overlap was found among the genes with intron retention defects in the three mutants (SuperExactTest, P < 0.05), indicating that MAC works as a complex in pre-mRNA splicing (Figure 8C). Two examples of intron retention events in these mutants are shown in Figure 8D.
Figure 8.
MAC Subunits Affect pre-mRNA Splicing.
(A) Box plot of intron/exon ratios in the indicated genotypes. All genes that pass an abundance filter in expression were included in this analysis. Asterisks indicate significant difference between the mutant and the wild type (Col-0) (Wilcoxon test, P < 2.2e-16).
(B) The intron/exon ratio per gene in Col-0 versus the indicated mutants. Black dots represent all genes with raw intronic read number ≥ 2, exonic read number ≥ 5, and final intron/exon ratio between 0 and 1. Red dots represent genes with significantly higher intron/exon ratio relative to the wild type (fold change ≥ 2 and false discovery rate < 0.01).
(C) A Venn diagram showing the degree of overlap among genes with intron retention defects in mac7-1, mac3a mac3b, and prl1 prl2.
(D) Examples of genes with intron retention defects. RPKM, reads per kilobase per million mapped reads. Two biological replicates for each genotype are shown. The rectangles mark introns with higher retention in the mutants.
We next examined whether there was any correlation between splicing defects and gene expression. We compared the expression levels of genes with intron retention defects in each mutant versus the wild type. As a reference, all genes that passed a minimum intron read coverage filter (see Figure 8 legend) were analyzed. There was no correlation between intron retention defects and the status of their differential expression in the mac mutants. Like all analyzed genes, genes with intron retention defects were increased, reduced, or unchanged in expression levels in each mutant compared with the wild type (Figure 9A; Supplemental Data Set 5). In addition, we examined intron versus exon ratios in total genes, upregulated genes, and downregulated genes in each mutant. Both upregulated and downregulated genes showed significant intron retention as total genes (Wilcoxon test, P < 2.2e-16), again indicating that the intron versus exon ratio has no correlation with gene expression changes (Figure 9B; Supplemental Data Set 4).
Figure 9.
MAC Affects pre-mRNA Splicing and Gene Expression Independently.
(A) Volcano plots illustrating fold changes of gene expression levels in the indicated mutants compared with Col-0. Thresholds for fold change of 1.5 and P ≤ 0.01 are shown in the plots as gray dashed lines. Black dots and red dots represent the same genes as in Figure 8B.
(B) Box plots of intron/exon ratios in total, upregulated, and downregulated genes in the indicated genotypes. Only genes that pass an abundance filter (raw intronic read number ≥ 2 and exonic read number ≥ 5) were included in this analysis. Asterisks indicate significant difference between the mutant and the wild type (Col-0) (Wilcoxon test, P < 2.2e-16).
Given the intron retention defects and miRNA accumulation defects in the mac mutants, we asked whether intron retention in pri-miRNAs contributed to the miRNA accumulation defects. Many MIR genes were shown to have introns, which are spliced out in pri-miRNAs (Laubinger et al., 2008; Zhan et al., 2012; Zielezinski et al., 2015). We performed RT-PCR with intron-flanking primers to detect unspliced miRNA precursors, including pri-miR163, pri-miR156, pri-miR166, pri-miR168, and pri-miR172. Genomic DNA was amplified with the same sets of primers to indicate the sizes of intron-containing fragments. No intron retention was observed in these miRNA precursors in mac7-1 (Supplemental Figure 9).
Thus, intron retention and differential expression of genes (including MIR genes) are not linked. MAC seems to regulate gene expression and RNA splicing independently.
DISCUSSION
MAC7 is an evolutionarily conserved protein across eukaryotes; however, little is known about the molecular and biological functions of MAC7 or its orthologs. The human ortholog Aquarius is an RNA helicase with ATPase activity, and it binds to introns to assist intron splicing in vitro (Hirose et al., 2006; De et al., 2015). The C. elegans ortholog EMB-4 was reported to act in the nuclear RNAi pathway, where it interacts with nuclear AGOs and functions in germline-specific chromatin remodeling (Checchi and Kelly, 2006; Akay et al., 2017; Tyc et al., 2017). In Arabidopsis, MAC7 is involved in plant defense and was predicted to be an essential gene (Tzafrir et al., 2004; Monaghan et al., 2009). In this study, we showed that a T-DNA insertion mutant of MAC7 is indeed embryo lethal, and we isolated the first viable mac7 mutant, mac7-1, as a miRNA biogenesis-defective mutant. In this mutant, the artificial miRNA, amiR-SUL, and many endogenous miRNAs show reduced accumulation. MAC7 was identified as a MAC subunit through MOS4 IP-MS (Monaghan et al., 2009); our MAC7 IP-MS confirmed this. Two other MAC subunits, CDC5 and PRL1, were previously shown to promote miRNA biogenesis (Zhang et al., 2013b, 2014). In this study, we showed that MAC3 also has a similar role. Thus, it is likely that MAC as a complex promotes miRNA biogenesis in general. Based on small RNA-seq analysis, MAC may promote the biogenesis of not only miRNAs but also siRNAs. Both 21- and 24-nucleotide small RNAs, the two most abundant small RNA size classes in Arabidopsis, are reduced in mac7-1 and prl1 prl2 mutants. A role of MAC in promoting siRNA biogenesis is also supported by previous findings on CDC5, MAC3, and PRL1 (Zhang et al., 2013a; Zhang et al., 2013b, 2014).
How does MAC7 promote miRNA biogenesis? In mac7-1, the reduced accumulation of miRNAs correlated with reduced levels of pri-miRNAs. Thus, MAC7 probably acts in miRNA biogenesis by promoting MIR transcription or pri-miRNA stability. However, the activity of a promoter driving MIR expression was not affected in mac7-1, nor was the endogenous SUC2 promoter activity. The half-lives of pri-miRNAs were not affected either. Although we cannot exclude the possibility that MAC7 promotes pri-miRNA processing into pre-mRNA or mature miRNAs and prevents pri-miRNA decay at the same time, and therefore pri-miRNA half-lives appeared unchanged in the mutant, we prefer to hypothesize that MAC7 plays a role in transcription elongation and/or maturation of pri-miRNAs, considering the interactions between Pol II and MAC subunits CDC5 and PRL1, as well as the functions of yeast NTC in transcription elongation (Kuraoka et al., 2008; Chanarat and Sträßer, 2013; Zhang et al., 2013b, 2014). Given the role of MAC7 in pre-mRNA splicing discovered before and in this study (Xu et al., 2012; Zhang et al., 2013a), we considered the possibility that MAC7 acts in miRNA biogenesis by promoting the splicing of pri-miRNAs. However, we did not detect intron retention in several intron-containing pri-miRNAs in mac7-1, although the mature miRNAs from those pri-miRNAs are reduced. In addition, MIR genes without any introns (e.g., the pSUC2:amiR-SUL transgene, MIR159a, and MIR167a) (Szarzynska et al., 2009; de Felippes et al., 2011) were also affected in mac7-1. Thus, the miRNA biogenesis defects of mac7-1 could not be attributed to defects in splicing. Intriguingly, we observed that MAC7 affects HYL1 localization to dicing bodies. Little is known about how dicing bodies form. MAC7 may recruit the dicing complex to pri-miRNAs through protein-protein interactions to form dicing bodies. Alternatively, dicing complex proteins are recruited by pri-miRNAs to dicing bodies; therefore, the reduced number of dicing bodies in mac7-1 could be a consequence of reduced pri-miRNA levels in the mutant. We favor the second hypothesis because MAC7 plays a more general role in transcription and RNA metabolism (such as splicing), while the dicing complex acts more specifically on miRNA precursors and is presumably not recruited to other RNAs that MAC7 may also act on.
CDC5 and PRL1 were shown to interact with dicing complex proteins (e.g., DCL1 and SERRATE) in vivo through coimmunoprecipitation or BiFC analyses (Zhang et al., 2013b, 2014). We did not detect interactions between MAC7 and the dicing complex through MAC7 IP-MS or BiFC analyses. However, many MAC components including MAC7 were found in HYL1 IP-MS, which clearly indicates association between the dicing complex and MAC in vivo. The possible reasons for the inability of MAC7 to pull down the dicing complex proteins are as follows: (1) The interaction between MAC7 and the dicing complex is indirect or weak, and bridged through CDC5 or PRL1; (2) only a small portion of MAC7 proteins interacts with the dicing complex. The second hypothesis is consistent with MAC7 having broader functions beyond miRNA biogenesis. We hypothesize that most of HYL1 or the dicing complex is associated with MAC, but not the other way around, which explains the recovery of MAC in HYL1 IP-MS.
MAC has functions beyond miRNA biogenesis in Arabidopsis. In our RNA-seq analysis, stress response-related GO terms were significantly enriched in downregulated genes in three MAC subunit mutants. This is consistent with previous studies showing that many mac mutants are more susceptible to pathogen infection (Monaghan et al., 2009, 2010; Weihmann et al., 2012; Xu et al., 2012). Although the mac7-1 hypo-DEGs are enriched in defense-related GO terms, the genes show the smallest reduction magnitude compared with other mac mutants, which could be the reason why mac7-1 is not susceptible to P. syringae infection as prl1-2 and mac3a mac3b mutants are (Monaghan et al., 2009; Weihmann et al., 2012).
It has been suspected that MAC is also involved in splicing in plants, like its orthologs in human and yeast (Johnson et al., 2011; Koncz et al., 2012). Indeed, our RNA-seq analyses uncovered intron retention defects in three MAC subunit mutants, thus linking MAC with pre-mRNA splicing. However, there were no significant correlations between intron retention and changes in gene expression. Genes with intron retention were upregulated, downregulated, or unchanged in the mac mutants. We also did not detect intron retention for intron-containing pri-miRNAs in mac7-1. It is likely that MAC has separate roles in RNA splicing and gene expression regulation (including the regulation of MIR genes). Thus, MAC has broad functions in nuclear RNA metabolism.
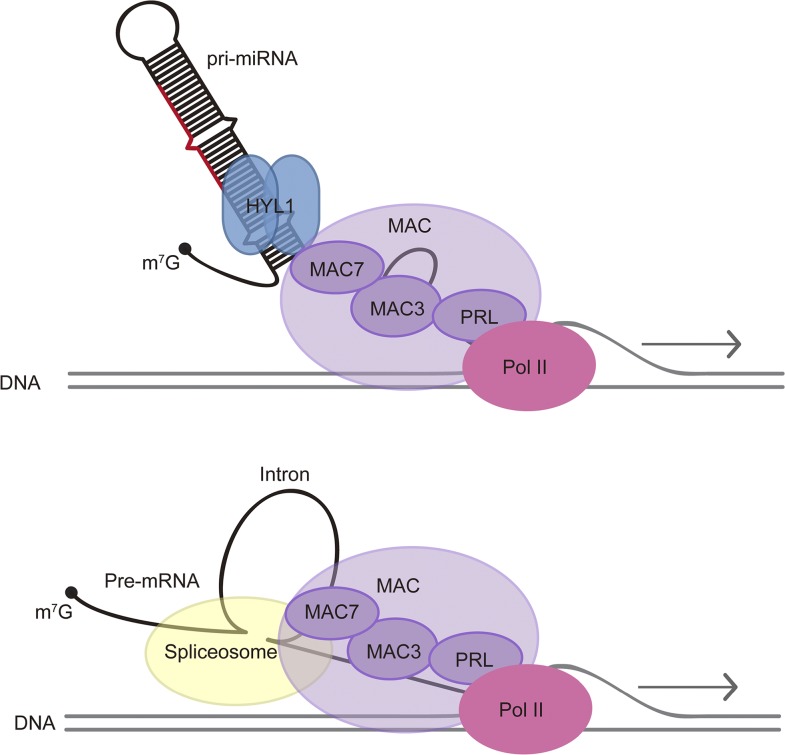
We speculate that a common theme of MAC’s role in nuclear RNA metabolism is linking RNA processing to transcription (Figure 10). In yeast, the NTC promotes transcription elongation (Chanarat et al., 2011; Chanarat and Sträßer, 2013). In both yeast and human, NTC or Prp19C associates with spliceosomes in vivo, although this association may entail NTC subcomplexes in human (Chanarat et al., 2012; De et al., 2015; Yan et al., 2015). In Arabidopsis, two MAC subunits, CDC5 and PRL1, have been shown to interact with Pol II in vivo (Zhang et al., 2013b, 2014). IP-MS of an Arabidopsis spliceosome subunit also pulled down multiple subunits of MAC (Deng et al., 2016). Thus, it is possible that MAC promotes cotranscriptional splicing through its interactions with both Pol II and the spliceosome. Similarly, in miRNA biogenesis, MAC may promote cotranscriptional pri-miRNA processing through its interactions with both Pol II and HYL1.
Figure 10.
A Model for MAC’s Functions in miRNA Biogenesis and pre-mRNA Processing.
MAC affects miRNA biogenesis through influencing Pol II transcription and interacting with the pri-miRNA processing factor HYL1. MAC also plays a role in pre-mRNA splicing through interactions with the spliceosome. MAC seems to have separate roles in miRNA biogenesis and RNA splicing, but a common theme appears to be cotranscriptional RNA processing.
METHODS
Plant Materials and Growth Conditions
The pSUC2:amiR-SUL transgenic line is a gift from Detlef Weigel (de Felippes et al., 2011). mac7-1 is a new allele isolated from our EMS mutagenesis screen with the pSUC2:amiR-SUL transgenic line. SALK_120944 (mac7-2) is a T-DNA insertion line obtained from the ABRC. The following published transgenic lines or mutants were used: pMIR167a:GUS and prl1-2 (SALK_008466) (Zhang et al., 2013b, 2014); p35S:HYL1-YFP (Qiao et al., 2015); p35S:YFP (Zhang et al., 2014); mac3a mac3b (mac3a is SALK_089300; mac3b is SALK_050811); and prl1 prl2 (prl1 is SALK_008466; prl2 is SALK_075970) double mutants (Monaghan et al., 2009; Weihmann et al., 2012).
In pMIR167a:GUS or p35S:HYL1-YFP transgenic lines, the transgene was confirmed to be homozygous by Basta selection (25 mg/L phosphinothricin; Gold Biotechnology) and by examining YFP signals under fluorescence microscopy, respectively, in multiple individuals. The mac7-1 mutant was crossed into pMIR167a:GUS or p35S:HYL1-YFP, and F2 plants containing homozygous transgenes were confirmed in the F3 generation.
Genotyping primers used are listed in Supplemental Table 4. Plants were grown in a plant growth chamber at 23°C for 16 h light (cool white fluorescent lamps, 25-W Sylvania 21942 FO25/741/ECO T8 linear tube) and 8-h dark cycles.
Mutagenesis Screening and Mapping of MAC7
The amiR-SUL mac7-1 M2 mutant was backcrossed with the parental line pSUC2:amiR-SUL. Genomic DNA was extracted from 100 pooled F2 mutants and used in library construction. The library was paired-end sequenced on Illumina’s HiSeq 2000 at ∼30× coverage, and the reads were mapped to the TAIR 10 genome using the Burrows-Wheeler Alignment tool (Li and Durbin, 2009). SamTools (Li et al., 2009) was used to identify EMS-induced single nucleotide polymorphisms. The single nucleotide polymorphism calls generated by SamTools were processed by the Next-Generation EMS mutation mapping website tools (Li et al., 2008). Two candidate mutations with 100% mutation rate were identified as EMS-typical C:G-to-T:A transitions that are predicted to cause nonsynonymous substitutions in the coding region of genes. Because one candidate mutation was only supported by three reads, we focused on the one in At2g38770 (MAC7/EMB2765), which was supported by 29 reads. A derived cleaved amplified polymorphic sequence (dCAPS) marker was designed to genotype this mutation (see Supplemental Table 4 for primers). The PCR products from the wild type can be digested by EcoRI, whereas those from mac7-1 could not. Linkage analysis was performed on 84 individual mutant plants in the F2 population of the backcross using this dCAPS marker to assess linkage between the mutation and the mac7-1 mutant phenotype.
DNA Constructs and Complementation
The MAC7 genomic region without the stop codon was amplified and cloned into pENTR/D-TOPO (Invitrogen) and then introduced to a modified pEarleyGate 301 vector (Earley et al., 2006) to generate pMAC7:MAC7-mCherry via LR reaction. The MAC7 genomic region without the stop codon was amplified and cloned into the pMDC107 gateway vector (Curtis and Grossniklaus, 2003) to generate pMAC7:MAC7-GFP with the Clontech In-Fusion HD Cloning Kit. The MAC7 coding region was cloned and then introduced into the pEarleyGate104 vector (Earley et al., 2006) via pENTR/D-TOPO (Invitrogen) through LR reactions. The above plasmids were used to transform mac7-1 plants through the Agrobacterium tumefaciens-mediated floral dip method. Primers used are listed in Supplemental Table 4.
Small RNA Gel Blotting, RT-PCR, and Quantitative RT-PCR
Total RNA from 2- to 3-week old seedlings (aerial part) or inflorescences was extracted with TRI reagent (Molecular Research Center). RNA gel blotting for detection of miRNAs was performed as described (Pall and Hamilton, 2008). Ten micrograms of total RNA from aerial part of seedlings or inflorescences was used in RNA gel blotting. The 5′ end 32P-labeled antisense DNA oligonucleotides were used to detect miRNAs. Oligonucleotide probes used are listed in Supplemental Table 4.
To perform RT-PCR, total RNA was first treated with DNase I (Roche) followed by reverse transcription using RevertAid reverse transcriptase (Thermo Fisher Scientific) with oligo-d(T) primers according to manufacturer’s instructions. Quantitative RT-PCR was performed in triplicate using iQ SYBRGreen Supermix (Bio-Rad) on the Bio-Rad CFX96 system. Primers used are listed in Supplemental Table 4.
RNA Half-Life Measurements
RNA half-life measurements were performed as described (Lidder et al., 2005) with minor modifications. Twelve-day-old Col and mac7-1 whole seedlings were transferred from Murashige and Skoog (MS) medium (PhytoTechnology Lab) agar plates to six-well plates with 0.5× MS medium and incubated overnight. The next day, cordycepin (Sigma-Aldrich) was added to a final concentration of 0.6 mM and the seedlings were collected for RNA extraction at 0, 30, 90, and 120 min after cordycepin addition. RT-qPCR was then performed to determine pri-miRNA levels. UBQ5 was used as an internal control.
Small RNA-Seq Library Construction and Data Analysis
To construct small RNA libraries, the aerial parts of 12-d-old Col-0, mac7-1, and prl1 prl2 seedlings grown on plates were harvested for total RNA extraction. Two biological replicates were included: Plants grown on different plates under the same conditions were collected at the same time into two separate samples for RNA extraction and subsequent procedures. To isolate small RNAs from total RNA, 50 µg of total RNA from each sample was resolved on 15% urea-PAGE gel, and the 18- to 30-nucleotide region was excised from the gel. Small RNAs were recovered by soaking the smashed gel in 0.3 M NaCl overnight, followed by ethanol precipitation. Small RNA libraries were constructed following instructions from the NEBNext Multiplex Small RNA Library Prep Set for Illumina (E7300). The libraries were sequenced on an Illumina HiSeq 2500 at the UC Riverside Genomics core facility.
Reads from small RNA-seq were first processed to remove the adaptor sequences by cutadapt (sequence: AGATCGGAA) (Martin, 2011). The reads were mapped to the TAIR10 genome using ShortStack with default parameters (Johnson et al., 2016). Normalization was performed by calculating the RPMR (reads per million of 45S rRNA reads) value (Li et al., 2016). Only one biological replicate for Col-0 was included in this analysis since the other Col-0 sample had very few reads caused by unknown problems in library construction or sequencing.
RNA-Seq Library Construction and Data Analysis
Polyadenylated RNA was isolated from total RNA extracted from 12-d-old seedlings (aerial part) of Col-0, mac7-1, mac3a mac3b, and prl1 prl2 using the Magnetic mRNA Isolation Kit (New England Biolabs), with two biological replicates for each genotype. For the biological replicates, plants grown on different plates under the same conditions were collected at the same time into two separate samples for RNA extraction and subsequent procedures. RNA-seq libraries were prepared using NEBNext mRNA Library Prep Reagent Set for Illumina (New England Biolabs) and sequenced on an Illumina HiSeq 2500 platform to generate high-quality single-end reads of 101 bp in length. Data analysis was performed with the pRNASeqTools pipeline (https://github.com/grubbybio/RNASeqTools). First, the raw reads were aligned to the TAIR10 genome using HISAT2 (Kim et al., 2015). Second, exonic and intronic reads were classified and quantified as follows. Reads with five or more nucleotides overlapping with intronic regions (intronic regions defined in all isoforms, including splicing donor/acceptor sites) were counted as intronic reads, and reads that mapped exclusively to exonic regions were counted as exonic reads. Transcript levels were measured in reads per million total read counts. Differentially expressed genes were identified using DEseq2 with fold change of 1.5 and P < 0.01 as the parameters (Love et al., 2014). To identify genes that exhibited significantly higher levels of intron retention compared with Col-0, the intron/exon ratio was calculated as (I1 + I2)/2 over (E1+E2)/2 (I, intronic reads; E, exonic reads; 1, biological replicate 1; 2, biological replicate 2), while applying an abundance cutoff (raw intronic read number ≥ 2 and exonic read number ≥ 5). We only considered intron/exon (I/E) ratios between 0 and 1. The genes with intron retention were identified using DEseq2 based on values of (Imutant/Emutant)/(ICol-0/E Col-0) and fold changes ≥ 2 and false discovery rate < 0.01 as parameters (Love et al., 2014). SuperExactTest was employed to access the statistical significance of DEGs or intron retention gene overlaps among mac7-1, mac3a mac3b, and prl1 prl2 (Wang et al., 2015).
Protein Sequence Alignment
Database searching of MAC7 homologs was performed at National Center for Biotechnology Information (NCBI) (www.ncbi.nlm.nih.gov/). Alignment of protein sequence was performed with Muscle (Edgar, 2004) and the alignments were edited with Jalview (Waterhouse et al., 2009).
Antibody Generation and Immunoblotting
To generate anti-MAC7 antibodies, a 5′ portion of the coding region of MAC7 corresponding to the first 416 amino acids of the protein was amplified (primers listed in Supplemental Table 4) and inserted into pMCSG7-His-MBP and pSUMO-His vectors, respectively. The constructs were then transformed into the Escherichia coli strain BL21 for protein expression. The recombinant proteins were purified by AKTA fast protein liquid chromatography (GE Healthcare) using the MBP-Trap or His-Trap column. The purified MBP-tagged protein was used as antigens to raise polyclonal antibodies in rabbits as described (Peterson et al., 2010). The antiserum was affinity-purified using a MAC7-SUMO-His-conjugated column. The purified antibodies were used in immunoblotting and IP-MS experiments. A similar approach was employed to generate the anti-CH42/SUL antibody. The full-length CH42 protein fused with SUMO-His tag was expressed and purified to immunize two rabbits. Affinity purified antibodies were used in immunoblotting analysis.
Other antibodies used in immunoblotting experiments include anti-GFP (Roche; catalog no. 11814460001), anti-AGO1 (Agrisera; AS09 527), anti-HYL1 (Agrisera; AS06 136), anti-SERRATE (Agrisera; AS06 136), anti-DCL1 (Agrisera; AS12 2102), and antitubulin (Sigma-Aldrich; T9026).
Proteomic Analysis
Total proteins from 12-d-old seedlings were extracted and immunoprecipitated with indicated antibodies. The IP products were resolved in SDS-PAGE. The antibody bands were removed and the samples were subjected to mass spectrometry as described (Sleat et al., 2006; Deng et al., 2016). Two biological replicated were performed, and the identified interacting proteins were those represented by peptides with high hits from both biological replicates.
Transient Expression of Fluorescent Fusion Proteins in Tobacco Leaf Epidermal Cells
The CDS of HYL1 was amplified and cloned into pENTR/D-TOPO (Invitrogen) and then introduced into the pGWB661 gateway vector (Nakamura et al., 2010) to generate p35S:TagRFP-HYL1. p35S:DCL1-YFP in the pEG101 vector was from a published study (Zhang et al., 2013b). The generation of p35S:YFP-MAC7 was described above. Agrobacterium GV3101::mp90 transformed with p35S:YFP-MAC7, p35S:DCL1-YFP, or p35S:TagRFP-HYL1 was used to infiltrate tobacco (Nicotiana benthamiana) leaves as described (Sparkes et al., 2006). The expression of fluorescent fusion proteins was observed using a Leica SP5 confocal laser scanning microscope.
BiFC Analysis
BiFC analysis was performed as described (Walter et al., 2004). Paired cCFP and nVenus constructs were coinfiltrated into N. benthamiana leaves. After 48 h, YFP signals and chlorophyll autofluorescence signals were excited at 488 nm and detected by Olympus Fluoview 500 confocal microscopy with a narrow band-pass filter (BA 505–525 nm).
Histochemical GUS Assay and Alexander’s Staining of Pollen
GUS staining was performed as described (Kim et al., 2011). Briefly, 12-d-old seedlings from Col-0 and mac7-1 harboring a homozygous pMIR167a:GUS transgene were vacuum infiltrated for 10 min and then incubated in GUS staining solution at 37°C for several hours until blue color became visible. Tissue clearing was performed with 70% ethanol for 1 to 2 d before imaging.
Alexander’s staining of pollen was performed as described (Peterson et al., 2010). The stained pollen grains were observed under a microscope equipped with a CCD camera (Olympus).
Accession Numbers
Genes referred to in this study correspond to the following Arabidopsis Genome Initiative locus identifiers: MAC7/EMB2765, AT2G38770; SUL/CHLORINA42, AT4G18480; MICRORNA156A, AT2G25095; MICRORNA159A, AT1G73687; MICRORNA163, AT1G66725; MICRORNA166A, AT2G46685; MICRORNA167A, AT3G22886; MICRORNA168A, AT4G19395; MICRORNA171A, AT3G51375; MICRORNA172A, AT2G28056; MICRORNA390B, AT5G58465; MICRORNA394B, AT1G76135; MICRORNA396B, AT5G35407; MICRORNA397A, AT4G05105; ACTIN8, AT1G49240; UBIQUITIN5, AT3G62250; TUBULIN3, AT5G62700; EIF4A1, AT3G13920; IPP2, AT3G02780 ;CDC5, AT1G09770; PRL1, AT4G15900; PRL2, AT3G16650; MAC3A, AT1G04510; MAC3B, AT2G33340 ; HYL1, AT1G09700; DCL1, AT1G01040; SERRATE, AT2G27100; AGO1, AT1G48410; HEN1, AT4G20910; MYB33 AT5G06100; SCL6-IV AT4G00150; SPL10 AT1G27370; MYB65, AT3G11440; ARF17, AT1G77850; ARF8, AT5G37020; SCL6-III At3g60630; and CUC2 AT5G53950. Protein sequences of MAC7 homologs in other species correspond to the following NCBI references: Homo sapiens intron binding protein aquarius, NP_055506.1; Mus musculus intron binding protein aquarius, NP_033832.2; Danio rerio intron binding protein aquarius, NP_956758; Drosophila melanogaster CG31368, NP_996198.2; Caenorhabditis elegans EMB-4, NP_001256831.1; and Schizosaccharomyces pombe Cwf11, NP_595360.1. RNA sequencing data are available from the NCBI Gene Expression Omnibus under the following reference numbers: Col-0_1, GSM2585832; Col-0_2, GSM2585833; mac7-1_1, GSM2585834; mac7-1_2, GSM2585835; mac3amac3b_1, GSM2585836; mac3amac3b_2, GSM2585837; prl1prl2_1, GSM2585838; prl1prl2_2, GSM2585839; Col-0_1_sRNA, GSM2771029; mac7-1_1_sRNA, GSM2771030; mac7-1_2_sRNA, GSM2771031; prl1 prl2_1_sRNA, GSM2771032; and prl1 prl2_2_sRNA, GSM2771033.
Supplemental Data
Supplemental Figure 1. The mac7-1 mutant shows pleiotropic developmental phenotypes.
Supplemental Figure 2. Reduced miRNA accumulation in both seedlings and inflorescences in the mac7-1 mutant.
Supplemental Figure 3. The mac7-1 mutation in the Col-0 background does not affect CH42 expression at either mRNA or protein levels.
Supplemental Figure 4. The MAC7 T-DNA insertion line, mac7-2, is a single-locus, recessive embryo-lethal mutant.
Supplemental Figure 5. Amino acid sequence alignment of Arabidopsis MAC7 orthologs.
Supplemental Figure 6. MAC7 does not affect the expression of the endogenous SUC2 gene or key miRNA biogenesis factors.
Supplemental Figure 7. MAC7 shows dispersed distribution in the nucleoplasm, while DCL1 and HYL1 concentrate in dicing bodies in the nuclei.
Supplemental Figure 8. Growth of virulent P.s.m. ES4326 at 0 and 3 days postinoculation.
Supplemental Figure 9. Intron-containing pri-miRNAs show similar splicing patterns in Col-0 and mac7-1.
Supplemental Table 1. Phenotypic and genotypic segregation in the progeny of selfed heterozygous mac7-2 plants.
Supplemental Table 2. MAC7-associated proteins identified by immunoprecipitation followed by mass spectrometry.
Supplemental Table 3. HYL1-associated MAC subunits identified by immunoprecipitation followed by mass spectrometry.
Supplemental Table 4. Oligonucleotide sequences.
Supplemental Data Set 1. Levels of 21-nt small RNAs, 24-nt small RNAs, and miRNAs in log2(reads per million of 45S rRNA reads) in Col-0, mac7-1, and prl1 prl2 as determined by small RNA-seq.
Supplemental Data Set 2. Differentially expressed genes in mac7-1, mac3a mac3b, and prl1 prl2 mutants as determined by RNA-seq analysis.
Supplemental Data Set 3. Gene Ontology analysis of overlapped DEGs among mac7-1, mac3a mac3b, and prl1 prl2 mutants.
Supplemental Data Set 4. Intron/exon ratio in Col-0, mac7-1, mac3a mac3b, and prl1 prl2.
Supplemental Data Set 5. Relative expression levels of genes that passed the intronic and exonic read counts filter in mac7-1, mac3a mac3b, and prl1 prl2 compared with Col-0, and relative expression levels of genes with intron retention defects in indicated mutants compared with Col-0.
Acknowledgments
We thank Detlef Weigel and Yongli Qiao for sharing the pSUC2:amiR-SUL and HYL1-YFP transgenic lines, respectively. We thank Fedor V. Karginov for comments and suggestions on this project, Brandon Le and Lorena Arroyo for technical advice and support, and Haiyan Zheng for help with mass spectrometry analysis. We thank Brandon Le, Shaofang Li, and Yu Yu for comments on this manuscript. We thank Li Liu and Wenrong He for the EMS treatment and seed planting, Jianbo Song for help with small RNA-seq analysis, and ZhongShou Wu for doing additional pathogen infection assays for article revision. This work was funded by the National Institutes of Health (GM061146) and Gordon and Betty Moore Foundation (GBMF3046) to X.C.
AUTHOR CONTRIBUTIONS
T.J. and X.C. designed the experiments. T.J., B.Z., S.L., Y.Z., K.C.M.J., B.Y., and X.L. performed the experiments. T.J., C.Y., and L.Z. analyzed the data. T.J., C.Y., and X.C. wrote the manuscript.
References
- Achkar N.P., Cambiagno D.A., Manavella P.A. (2016). miRNA biogenesis: A dynamic pathway. Trends Plant Sci. 21: 1034–1044. [DOI] [PubMed] [Google Scholar]
- Akay A., et al. (2017). The helicase Aquarius/EMB-4 is required to overcome intronic barriers to allow nuclear RNAi pathways to heritably silence transcription. Dev. Cell 42: 241–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribas-Hernández L., Marchais A., Poulsen C., Haase B., Hauptmann J., Benes V., Meister G., Brodersen P. (2016). The slicer activity of ARGONAUTE1 is required specifically for the phasing, not production, of trans-acting short interfering RNAs in Arabidopsis. Plant Cell 28: 1563–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N., Baulcombe D.C. (2005). Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA 102: 11928–11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell A., Fahlgren N., Garcia-Ruiz H., Gilbert K.B., Montgomery T.A., Nguyen T., Cuperus J.T., Carrington J.C. (2012). Functional analysis of three Arabidopsis ARGONAUTES using slicer-defective mutants. Plant Cell 24: 3613–3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanarat S., Sträßer K. (2013). Splicing and beyond: the many faces of the Prp19 complex. Biochim. Biophys. Acta 1833: 2126–2134. [DOI] [PubMed] [Google Scholar]
- Chanarat S., Seizl M., Strässer K. (2011). The Prp19 complex is a novel transcription elongation factor required for TREX occupancy at transcribed genes. Genes Dev. 25: 1147–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanarat S., Burkert-Kautzsch C., Meinel D.M., Sträßer K. (2012). Prp19C and TREX: interacting to promote transcription elongation and mRNA export. Transcription 3: 8–12. [DOI] [PubMed] [Google Scholar]
- Checchi P.M., Kelly W.G. (2006). emb-4 is a conserved gene required for efficient germline-specific chromatin remodeling during Caenorhabditis elegans embryogenesis. Genetics 174: 1895–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De I., Bessonov S., Hofele R., dos Santos K., Will C.L., Urlaub H., Lührmann R., Pena V. (2015). The RNA helicase Aquarius exhibits structural adaptations mediating its recruitment to spliceosomes. Nat. Struct. Mol. Biol. 22: 138–144. [DOI] [PubMed] [Google Scholar]
- de Felippes F.F., Ott F., Weigel D. (2011). Comparative analysis of non-autonomous effects of tasiRNAs and miRNAs in Arabidopsis thaliana. Nucleic Acids Res. 39: 2880–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Lu T., Wang L., Gu L., Sun J., Kong X., Liu C., Cao X. (2016). Recruitment of the NineTeen Complex to the activated spliceosome requires AtPRMT5. Proc. Natl. Acad. Sci. USA 113: 5447–5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z., Han M.H., Fedoroff N. (2008). The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc. Natl. Acad. Sci. USA 105: 9970–9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X., Cui Y., Li Y., Qi Y. (2015). Transcription and processing of primary microRNAs are coupled by Elongator complex in Arabidopsis. Nat. Plants 1: 15075. [DOI] [PubMed] [Google Scholar]
- Fang Y., Spector D.L. (2007). Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr. Biol. 17: 818–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco-Mangilet A.G., Karlsson P., Kim M.H., Eo H.J., Oh S.A., Kim J.H., Kulcheski F.R., Park S.K., Manavella P.A. (2015). THO2, a core member of the THO/TREX complex, is required for microRNA production in Arabidopsis. Plant J. 82: 1018–1029. [DOI] [PubMed] [Google Scholar]
- Furumizu C., Tsukaya H., Komeda Y. (2010). Characterization of EMU, the Arabidopsis homolog of the yeast THO complex member HPR1. RNA 16: 1809–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium (2015). Gene Ontology Consortium: going forward. Nucleic Acids Res. 43: D1049–D1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajheidari M., Farrona S., Huettel B., Koncz Z., Koncz C. (2012). CDKF;1 and CDKD protein kinases regulate phosphorylation of serine residues in the C-terminal domain of Arabidopsis RNA polymerase II. Plant Cell 24: 1626–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M.H., Goud S., Song L., Fedoroff N. (2004). The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl. Acad. Sci. USA 101: 1093–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T., Ideue T., Nagai M., Hagiwara M., Shu M.D., Steitz J.A. (2006). A spliceosomal intron binding protein, IBP160, links position-dependent assembly of intron-encoded box C/D snoRNP to pre-mRNA splicing. Mol. Cell 23: 673–684. [DOI] [PubMed] [Google Scholar]
- Ji L., et al. (2011). ARGONAUTE10 and ARGONAUTE1 regulate the termination of floral stem cells through two microRNAs in Arabidopsis. PLoS Genet. 7: e1001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K.C.M., Dong O.X., Li X. (2011). The evolutionarily conserved MOS4-associated complex. Cent. Eur. J. Biol. 6: 776–784. [Google Scholar]
- Johnson N.R., Yeoh J.M., Coruh C., Axtell M.J. (2016). Improved placement of multi-mapping small RNAs. G3 (Bethesda) 6: 2103–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson P., Christie M.D., Seymour D.K., Wang H., Wang X., Hagmann J., Kulcheski F., Manavella P.A. (2015). KH domain protein RCF3 is a tissue-biased regulator of the plant miRNA biogenesis cofactor HYL1. Proc. Natl. Acad. Sci. USA 112: 14096–14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S.L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.J., Zheng B., Yu Y., Won S.Y., Mo B., Chen X. (2011). The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J. 30: 814–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C., Dejong F., Villacorta N., Szakonyi D., Koncz Z. (2012). The spliceosome-activating complex: molecular mechanisms underlying the function of a pleiotropic regulator. Front. Plant Sci. 3: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraoka I., et al. (2008). Isolation of XAB2 complex involved in pre-mRNA splicing, transcription, and transcription-coupled repair. J. Biol. Chem. 283: 940–950. [DOI] [PubMed] [Google Scholar]
- Kurihara Y., Watanabe Y. (2004). Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. USA 101: 12753–12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y., Takashi Y., Watanabe Y. (2006). The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA 12: 206–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S., Sachsenberg T., Zeller G., Busch W., Lohmann J.U., Rätsch G., Weigel D. (2008). Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 105: 8795–8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Ruan J., Durbin R. (2008). Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18: 1851–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Yang Z., Yu B., Liu J., Chen X. (2005). Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol. 15: 1501–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., et al. (2016). Biogenesis of phased siRNAs on membrane-bound polysomes in Arabidopsis. eLife pii: e22750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidder P., Gutiérrez R.A., Salomé P.A., McClung C.R., Green P.J. (2005). Circadian control of messenger RNA stability. Association with a sequence-specific messenger RNA decay pathway. Plant Physiol. 138: 2374–2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z., Yin K., Zhu D., Chen Z., Gu H., Qu L.J. (2007). AtCDC5 regulates the G2 to M transition of the cell cycle and is critical for the function of Arabidopsis shoot apical meristem. Cell Res. 17: 815–828. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavella P.A., Hagmann J., Ott F., Laubinger S., Franz M., Macek B., Weigel D. (2012). Fast-forward genetics identifies plant CPL phosphatases as regulators of miRNA processing factor HYL1. Cell 151: 859–870. [DOI] [PubMed] [Google Scholar]
- Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10–12. [Google Scholar]
- Monaghan J., Xu F., Xu S., Zhang Y., Li X. (2010). Two putative RNA-binding proteins function with unequal genetic redundancy in the MOS4-associated complex. Plant Physiol. 154: 1783–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan J., Xu F., Gao M., Zhao Q., Palma K., Long C., Chen S., Zhang Y., Li X. (2009). Two Prp19-like U-box proteins in the MOS4-associated complex play redundant roles in plant innate immunity. PLoS Pathog. 5: e1000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S., Mano S., Tanaka Y., Ohnishi M., Nakamori C., Araki M., Niwa T., Nishimura M., Kaminaka H., Nakagawa T., Sato Y., Ishiguro S. (2010). Gateway binary vectors with the bialaphos resistance gene, bar, as a selection marker for plant transformation. Biosci. Biotechnol. Biochem. 74: 1315–1319. [DOI] [PubMed] [Google Scholar]
- Németh K., et al. (1998). Pleiotropic control of glucose and hormone responses by PRL1, a nuclear WD protein, in Arabidopsis. Genes Dev. 12: 3059–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ner-Gaon H., Halachmi R., Savaldi-Goldstein S., Rubin E., Ophir R., Fluhr R. (2004). Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J. 39: 877–885. [DOI] [PubMed] [Google Scholar]
- Ozgur S., Buchwald G., Falk S., Chakrabarti S., Prabu J.R., Conti E. (2015). The conformational plasticity of eukaryotic RNA-dependent ATPases. FEBS J. 282: 850–863. [DOI] [PubMed] [Google Scholar]
- Pall G.S., Hamilton A.J. (2008). Improved northern blot method for enhanced detection of small RNA. Nat. Protoc. 3: 1077–1084. [DOI] [PubMed] [Google Scholar]
- Palma K., Zhao Q., Cheng Y.T., Bi D., Monaghan J., Cheng W., Zhang Y., Li X. (2007). Regulation of plant innate immunity by three proteins in a complex conserved across the plant and animal kingdoms. Genes Dev. 21: 1484–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W., Li J., Song R., Messing J., Chen X. (2002). CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12: 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson R., Slovin J.P., Chen C. (2010). A simplified method for differential staining of aborted and non-aborted pollen grains. Int. J. Plant Biol. 1: 66–69. [Google Scholar]
- Qiao Y., Shi J., Zhai Y., Hou Y., Ma W. (2015). Phytophthora effector targets a novel component of small RNA pathway in plants to promote infection. Proc. Natl. Acad. Sci. USA 112: 5850–5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raczynska K.D., Stepien A., Kierzkowski D., Kalak M., Bajczyk M., McNicol J., Simpson C.G., Szweykowska-Kulinska Z., Brown J.W., Jarmolowski A. (2014). The SERRATE protein is involved in alternative splicing in Arabidopsis thaliana. Nucleic Acids Res. 42: 1224–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G., Xie M., Dou Y., Zhang S., Zhang C., Yu B. (2012). Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 12817–12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers K., Chen X. (2013). Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25: 2383–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleat D.E., Zheng H., Qian M., Lobel P. (2006). Identification of sites of mannose 6-phosphorylation on lysosomal proteins. Mol. Cell. Proteomics 5: 686–701. [DOI] [PubMed] [Google Scholar]
- Song L., Han M.H., Lesicka J., Fedoroff N. (2007). Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc. Natl. Acad. Sci. USA 104: 5437–5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I.A., Runions J., Kearns A., Hawes C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1: 2019–2025. [DOI] [PubMed] [Google Scholar]
- Szarzynska B., Sobkowiak L., Pant B.D., Balazadeh S., Scheible W.R., Mueller-Roeber B., Jarmolowski A., Szweykowska-Kulinska Z. (2009). Gene structures and processing of Arabidopsis thaliana HYL1-dependent pri-miRNAs. Nucleic Acids Res. 37: 3083–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyc K.M., Nabih A., Wu M.Z., Wedeles C.J., Sobotka J.A., Claycomb J.M. (2017). The conserved intron binding protein EMB-4 plays differential roles in germline small RNA pathways of C. elegans. Dev. Cell 42: 256–270. [DOI] [PubMed] [Google Scholar]
- Tzafrir I., Pena-Muralla R., Dickerman A., Berg M., Rogers R., Hutchens S., Sweeney T.C., McElver J., Aux G., Patton D., Meinke D. (2004). Identification of genes required for embryo development in Arabidopsis. Plant Physiol. 135: 1206–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438. [DOI] [PubMed] [Google Scholar]
- Wang L., Song X., Gu L., Li X., Cao S., Chu C., Cui X., Chen X., Cao X. (2013). NOT2 proteins promote polymerase II-dependent transcription and interact with multiple microRNA biogenesis factors in Arabidopsis. Plant Cell 25: 715–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Zhao Y., Zhang B. (2015). Efficient test and visualization of multi-set intersections. Sci. Rep. 5: 16923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A.M., Procter J.B., Martin D.M., Clamp M., Barton G.J. (2009). Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihmann T., Palma K., Nitta Y., Li X. (2012). Pleiotropic regulatory locus 2 exhibits unequal genetic redundancy with its homolog PRL1. Plant Cell Physiol. 53: 1617–1626. [DOI] [PubMed] [Google Scholar]
- Wiborg J., O’Shea C., Skriver K. (2008). Biochemical function of typical and variant Arabidopsis thaliana U-box E3 ubiquitin-protein ligases. Biochem. J. 413: 447–457. [DOI] [PubMed] [Google Scholar]
- Wu X., Shi Y., Li J., Xu L., Fang Y., Li X., Qi Y. (2013). A role for the RNA-binding protein MOS2 in microRNA maturation in Arabidopsis. Cell Res. 23: 645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Allen E., Fahlgren N., Calamar A., Givan S.A., Carrington J.C. (2005). Expression of Arabidopsis MIRNA genes. Plant Physiol. 138: 2145–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Xu S., Wiermer M., Zhang Y., Li X. (2012). The cyclin L homolog MOS12 and the MOS4-associated complex are required for the proper splicing of plant resistance genes. Plant J. 70: 916–928. [DOI] [PubMed] [Google Scholar]
- Yan C., Hang J., Wan R., Huang M., Wong C.C., Shi Y. (2015). Structure of a yeast spliceosome at 3.6-angstrom resolution. Science 349: 1182–1191. [DOI] [PubMed] [Google Scholar]
- Yang L., Liu Z., Lu F., Dong A., Huang H. (2006a). SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 47: 841–850. [DOI] [PubMed] [Google Scholar]
- Yang Z., Ebright Y.W., Yu B., Chen X. (2006b). HEN1 recognizes 21-24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res. 34: 667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Yang Z., Li J., Minakhina S., Yang M., Padgett R.W., Steward R., Chen X. (2005). Methylation as a crucial step in plant microRNA biogenesis. Science 307: 932–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Bi L., Zhai J., Agarwal M., Li S., Wu Q., Ding S.W., Meyers B.C., Vaucheret H., Chen X. (2010). siRNAs compete with miRNAs for methylation by HEN1 in Arabidopsis. Nucleic Acids Res. 38: 5844–5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Bi L., Zheng B., Ji L., Chevalier D., Agarwal M., Ramachandran V., Li W., Lagrange T., Walker J.C., Chen X. (2008). The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc. Natl. Acad. Sci. USA 105: 10073–10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X., Wang B., Li H., Liu R., Kalia R.K., Zhu J.K., Chinnusamy V. (2012). Arabidopsis proline-rich protein important for development and abiotic stress tolerance is involved in microRNA biogenesis. Proc. Natl. Acad. Sci. USA 109: 18198–18203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.J., Zhou J.X., Liu J., Ma Z.Y., Zhang S.W., Dou K., Huang H.W., Cai T., Liu R., Zhu J.K., He X.J. (2013a). The splicing machinery promotes RNA-directed DNA methylation and transcriptional silencing in Arabidopsis. EMBO J. 32: 1128–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Liu Y., Yu B. (2014). PRL1, an RNA-binding protein, positively regulates the accumulation of miRNAs and siRNAs in Arabidopsis. PLoS Genet. 10: e1004841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Xie M., Ren G., Yu B. (2013b). CDC5, a DNA binding protein, positively regulates posttranscriptional processing and/or transcription of primary microRNA transcripts. Proc. Natl. Acad. Sci. USA 110: 17588–17593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B., Wang Z., Li S., Yu B., Liu J.Y., Chen X. (2009). Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev. 23: 2850–2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielezinski A., et al. (2015). mirEX 2.0 - an integrated environment for expression profiling of plant microRNAs. BMC Plant Biol. 15: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]