Table 2. Diffraction Data Collection and Refinement Statistics.
| Data Set | 14-3-3c:CPP | 14-3-3c:CPP7 | 14-3-3c:CPP:FC |
|---|---|---|---|
| Data collection | |||
| Space group | P6522 | P6522 | P212121 |
| Cell dimensions | |||
| a, b, c (Å) | 110.18, 110.18, 136.69 | 109.03, 109.03, 136.71 | 99.81, 165.66, 170.55 |
| α, β, γ (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 90.0 |
| Wavelength (Å) | 0.96770 | 0.96770 | 0.97625 |
| Resolution (Å) | 47.71–2.35 (2.43–2.35) | 47.21–2.07 (2.13–2.07) | 48.32–3.30 (3.42–3.30) |
| #Rpim | 0.045 (0.365) | 0.037 (0.356) | 0.132 (0.304) |
| +CC1/2 | 0.971 (0.706) | 0.999 (0.523) | 0.978 (0.407) |
| <I/σ(I)> | 15.2 (5.1) | 18.3 (5.1) | 5.6 (2.7) |
| Completeness (%) | 100 (100) | 99.8 (99.8) | 100 (100) |
| Wilson B-factor | 32.28 | 29.44 | 45.76 |
| Redundancy | 39.0 (27.7) | 40.9 (39.8) | 7.5 (7.6) |
| Refinement | |||
| Resolution (Å) | 42.89–2.35 (2.43–2.35) | 44.63–2.07 (2.14–2.07) | 47.89-3.30 (3.41–3.30) |
| Number of reflections | 21,014 (2,044) | 29,779 (2,904) | 43,249 (4,259) |
| Rwork/Rfree | 0.195/0.245 (0.340/0.338) | 0.200/0.247 (0.327-0.357) | 0.241/0.306 (0.291/0.381) |
| Number of molecules | |||
| Copies in the AU | 1 | 1 | 8 |
| Protein residues | 245 | 247 | 1898 |
| FSC molecules | – | – | 7 |
| PEG molecules | – | 3 | – |
| GOL molecules | – | 1 | – |
| ACT molecules | 1 | 1 | – |
| Water molecules | 148 | 185 | 101 |
| Average B factors (Å2) | 51.5 | 46.3 | 63.4 |
| Average B factors (FSC) (Å2) | – | – | 49.9 |
| RMSD | |||
| Bond lengths (Å) | 0.013 | 0.015 | 0.002 |
| Bond angles (°) | 1.15 | 1.22 | 0.51 |
| Ramachandran plot statistics | 99% in favored | 98% in favored | 98% in favored |
| 0.0% outliers | 0.0% outliers | 0.0% outliers | |
Highest-resolution shell is shown in parentheses. +CC1/2 is the correlation coefficient of the mean intensities between two random half-sets of data. RMSD, root mean square deviation. 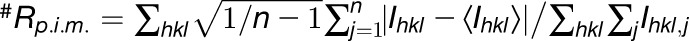 .
.
