Figure 1.
Identification of the SCD Complex and Its Interactors.
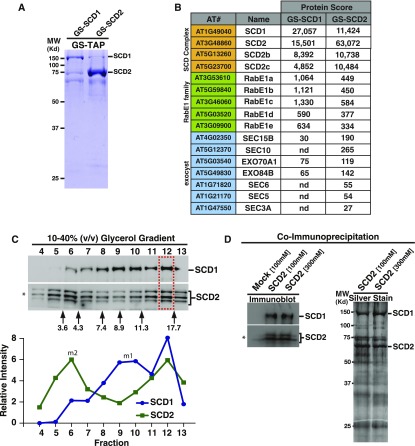
(A) Coomassie-stained SDS-PAGE analysis of TCA-precipitated TAP-purified GS-SCD1 and GS-SCD2 fractions.
(B) Abridged list of proteins that copurified with GS-SCD1 and GS-SCD2 as identified by LC/MS/MS. Shown is the Mascot protein score.
(C) Cofractionation of SCD1 and SCD2. Glycerol gradient velocity sedimentation analysis of native SCD1 and SCD2 from Arabidopsis cell extracts. Glycerol gradient fractions were analyzed by quantitative immunoblotting using indicated antibodies. Arrows indicate the fractionation of molecular mass standards with their indicated Svedberg (S)-values. Graph depicts the intensity of the SCD1 and SCD2 immunoblot signal peaks. SCD1 and SCD2 fractionate in separate lower molecular weight peaks corresponding to their expected monomeric molecular masses 132 and 64 kD, respectively (labeled m1 and m2), as well as in an overlapping (∼430 kD) peak (fraction 12; boxed region).
(D) Co-IP of SCD2 and SCD1. Immunoblot and silver stain analysis of Arabidopsis cell extracts immunoprecipitated without (mock) or with anti-SCD2 antibodies in the presence of 100 or 300 mM KCl. Asterisk: affinity-purified anti-SCD2 antibodies detect multiple SCD2 bands as described (McMichael et al., 2013).