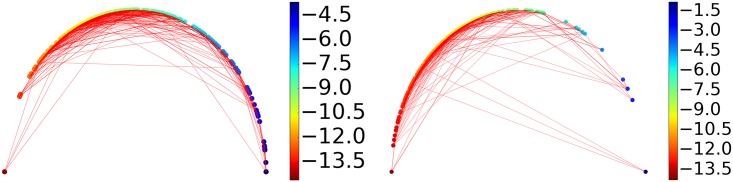
Fig 3. An illustration of similarities between ligands measured by their barcode space Wasserstein distances.
Ligands are ordered according to their binding affinities and are represented as dots on the semicircle. Specifically, a sample of binding free energy x is plotted at the angle θ = π(Emax − x)/(Emax − Emin) where Emin and Emax are the lowest and the highest energy in the dataset. Each dot is connected with two nearest neighbors based on their barcode space Wasserstein distances. An optimal prediction would be achieved if lines stay close to the semicircle. The majority of the connections stay near the boundary to the upper half sphere demonstrating that barcode space metric based Wasserstein distance measurement reflects the similarity in function, i.e., the binding affinity in this case. The protein clusters with the best and the worst performance are shown. Left: Protein cluster 2. Right: Protein cluster 3.