Figure 5. Genome-wide copy number variation in spontaneous and NNK-derived Gprc5a-/- LUADs.
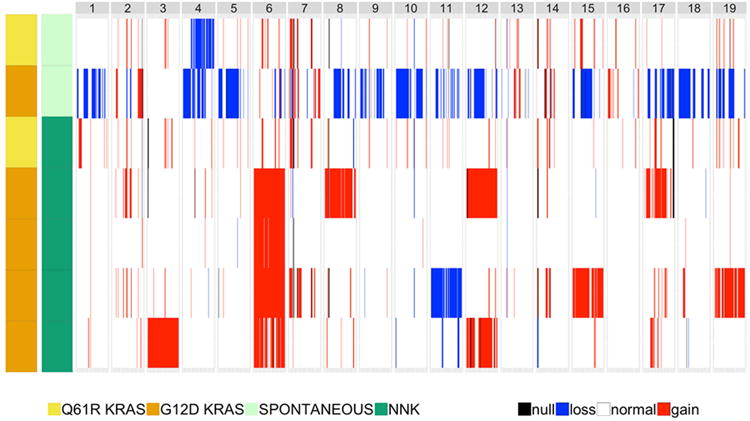
Copy number variations (CNVs) were assessed from sequencing data and read depth using FREEC and Sequenza as described in the Supplementary Methods. Read depth was analyzed in LUADs relative to depth in normal samples to estimate copy number in 8 kb windows followed by segmentation via a LASSO-based algorithm. Genome-wide CNAs across all LUADs were then plotted. Loss, blue; gain, red; not discernable, black.
