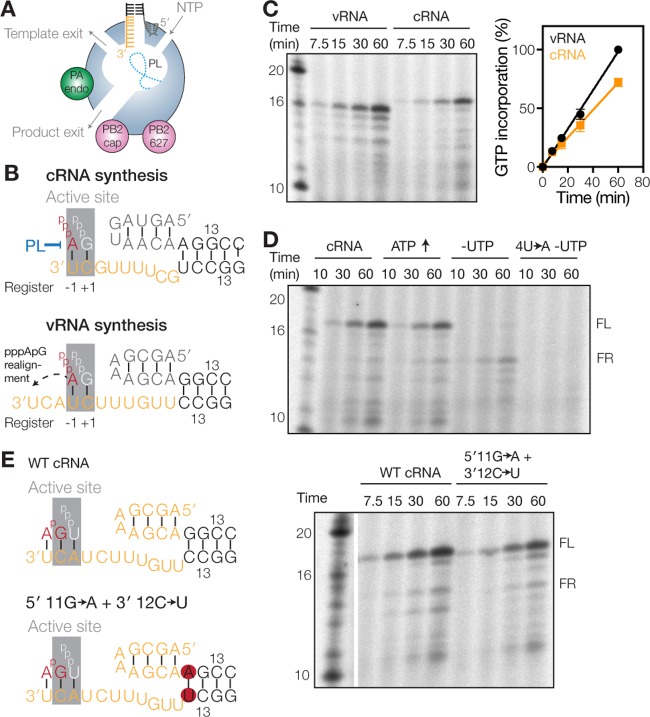
FIG 1.
Failed priming and realignment events can be detected in vitro. (A) Model of the influenza A virus RdRp. The core of the RdRp is shaded blue, the PA endonuclease (PA endo) is in green, and the PB2 cap-binding domain (PB2 cap) and 627 domain (PB2 627) are in pink. The priming loop (PL) is indicated as a dotted line. (B) Schematics of IAV initiation during cRNA and vRNA synthesis. The priming loop is indicated in blue. Active-site (gray) positions −1 and +1 are indicated below each schematic. (C) Time course of ApG extension on a vRNA or cRNA promoter. The graph shows the percentage of [α-32P]GTP incorporation relative to activity on the vRNA promoter. Error bars indicate standard deviations (n = 3). (D) Time course of ApG extension on a wild-type or 4U→A mutant cRNA promoter. The addition of 0.5 mM ATP or the omission of UTP is indicated. (E) Schematic of ApG extension on a wild-type (WT) cRNA promoter or a promoter where the first G-C base pair of the promoter duplex was mutated to A-U (5′ 11G→A+3′ 12C→U). The gel image shows results of an ApG extension assay on the wild-type cRNA promoter or the 5′ 11G→A+3′ 12C→U promoter mutant.