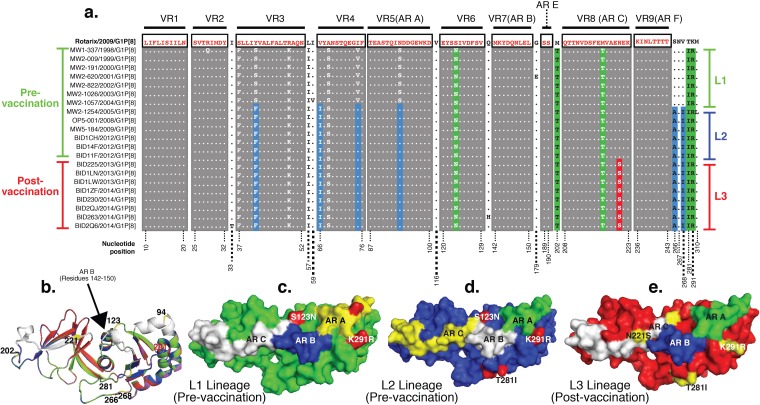
FIG 5.
Amino acid substitutions and structural conformation of the outer capsid glycoprotein of Malawian G1P[8] strains. (a) Complete VP7 sequences of representative pre- and postvaccine G1P[8] strains aligned to that of RV1 exhibiting amino acid substitutions that occurred within the variable regions (VR) and mapped antigenic regions (AR) over time. Lineage-defining amino acid amino acid substitutions are highlighted in green, blue, and yellow for the L1, L2, and L3 lineages, respectively. Pre- and postvaccine strains are shown with vertical green and red bars on the right, respectively. Strains belonging to the L1, L2, and L3 phylogenetic clusters are shown with green, blue, and red bars, respectively, on the right. (b) Perfect alignment of superimposed VP7 structures exhibiting few differences between RV1 and L1 to L3 strains. Antigenic regions A, B, and C are shown in white. L1 to L3 and RV1 strains are shown in yellow, green, blue, and red, respectively. (c to e) Surface visualization of VP7 from outside the virion on the 3-fold axis displaying amino acid differences when structures for L1 (c), L2 (d), and L3 (e) G1P[8] strains were superimposed on the structure of the outer capsid glycoprotein of RV1. Numbers correspond to the positions where mutations occurred.