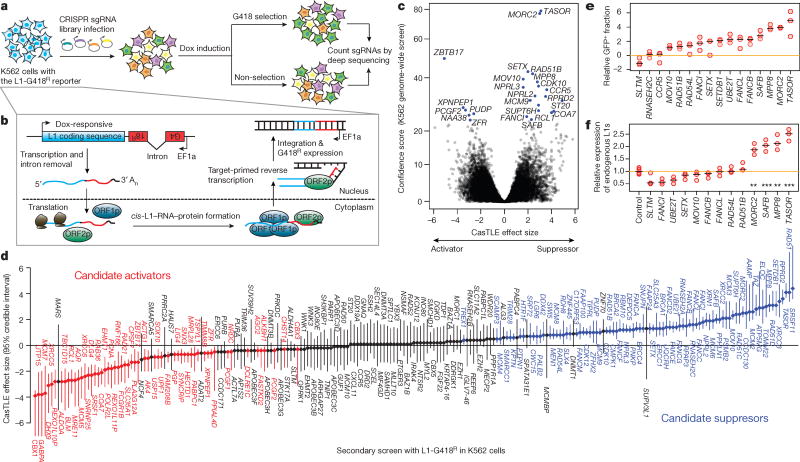
Figure 1.
Genome-wide screen for L1 activators and suppressors in K562 cells.
a. Schematic for the screen.
b. Schematic for the L1-G418R retrotransposition.
c. CasTLE analysis of (n = 2) independent K562 genome-wide screens. Genes at 10% FDR cutoff colored in blue, CasTLE likelihood ratio test11.
d. The maximum effect size (center value) estimated by CasTLE from two independent K562 secondary screens with 10 independent sgRNAs per gene. Bars, 95% credible interval (CI). L1 activators, red; L1 suppressors, blue; insignificant genes whose CI include 0, gray.
e. L1-GFP retrotransposition in control (infected with negative control sgRNAs, hereinafter referred to as ‘Ctrl’) and mutant K562 cells as indicated. GFP(+) cell fractions normalized to Ctrl. Center value as median. n = 3 biological replicates per gene.
f. RT-qPCR measuring endogenous L1Hs expression in mutant K562 cells, normalized to Ctrl. Center value as median. n = 3 technical replicates per gene. **P < 0.01; ***P < 0.001; two-sided Welch t-test.