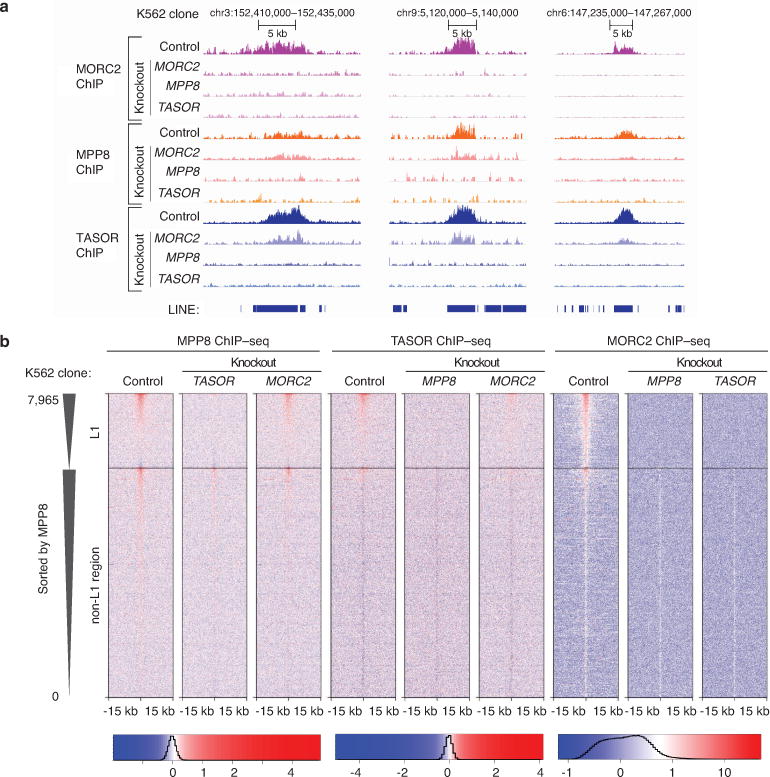
Extended Data Figure 7.
HUSH/MORC2 preferentially bind full-length L1 instances in human ESCs, mouse ESCs and K562 cells.
a. Widespread genomic co-binding of MPP8 and MORC2 in hESCs. Heatmap representation of ChIP-seq results at 57,000 genomic loci, centered on MPP8 and MORC2 summits and sorted by MORC2 ChIP-seq signal. Plotted is normalized ChIP read density from hESCs.
b. Heatmaps of MORC2/MPP8 ChIP-seq density over indicated repeat classes, centered and sorted as in panel a. HUSH complex and MORC2 bind predominantly to L1 elements in hESCs, in particular to the primate-specific L1P families, suggesting that HUSH/MORC2-dependent silencing is relevant in many embryonic and somatic cell types.
c. L1 families that encompass active L1 copies, such as L1Md-T and L1Md-A, are significantly enriched among MPP8 binding sites in mouse ESC. L1Md_Gf is also enriched but not shown due to the low number of instances. Thus, HUSH-mediated L1 regulation appears to be conserved among species. Of note, MPP8 is also strongly enriched at IAP elements, a class of murine endogenous retroviruses that remain currently mobile in the mouse genome.
d. MPP8 ChIP-seq heatmaps in mESCs featuring retrotransposition-competent L1Md-T, L1Md-A and L1Md-Gf.
e. MPP8 preferentially bind full-length L1Md-A and L1Md-T in mESCs. Plotted is size distribution of the indicated L1 instances that overlap with MPP8 ChIP-seq peaks, or remaining L1s that do not overlap with such ChIP-seq signals. Box plots show median and interquartile range (IQR), whiskers are 1.5× IQR.
f. Aggregate plots of MORC2 (red) and MPP8 (black) ChIP-seq signals over 500 full-length, MPP8-bound L1PAs, centered on the L1 5’ end.
g. Aggregate plots of MORC2 (red) and MPP8 (black) ChIP-seq signals on L1Hs (L1PA1). Similar as the binding profile on L1PA (panel f), MPP8/MORC2 occupy the whole body of L1Hs, with MORC2 additionally binding L1Hs 5′UTR. Please note that ChIP-seq fragments are much less likely to be uniquely mapped, and thus removed by the alignment criteria, within the L1Hs non-5’UTR region, due to their minimal sequence divergence (Extended Data Fig. 5a).