Figure 2. Definition and validation of CD1C+ DC subsets.
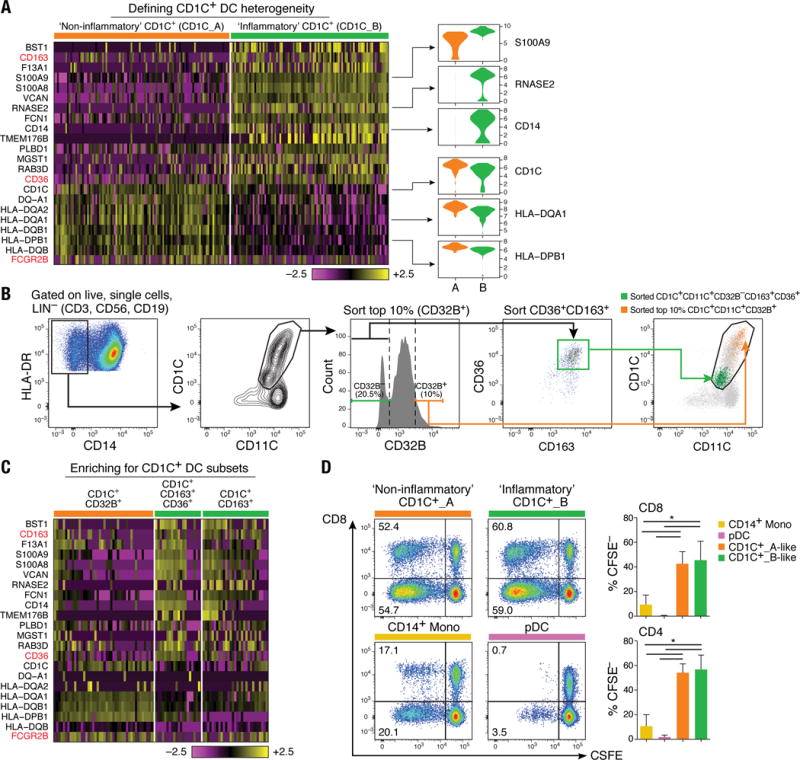
(A) Heatmap showing scaled expression (log TPM values) of discriminative gene sets defining each CD1C+ DC subset with AUC cutoff ≥ 0.75. Color scheme is based on z-score distribution, from −2.5 (purple) to 2.5 (yellow). Violin plots illustrate expression distribution of candidate genes across subsets on the x-axis (orange for CD1C_A/DC2; green for CD1C_B/DC3). In red are three markers used for subsequent enrichment strategy: CD163, CD36 and FCGR2B/CD32B (AUC =0.63). (B) Enrichment gating strategy of CD1C+ DC subsets [LIN(CD3, CD19, CD56)−HLA-DR+CD14−CD1C+CD11C+]. The CD1C_A/DC2 subset was further enriched by sorting on the 10% brightest CD32B+ cells (orange gate); the CD1C_B/DC3 subset was enriched by sorting on CD32B−CD163+CD36+ cells (green gate) or on CD32B−CD163+. Right: Overlay of the two sorted CD1C+ DC populations; 47 single cells were sorted from the green and orange gates in a 96-well plate for profiling. (C) Heatmap reporting scaled expression (log TPM values) of scRNAseq data from three cell subsets defined by CD1C+CD32B+, CD1C+CD36+CD163+, and CD1C+CD163+. Either CD1C+CD36+CD163+ or CD1C+CD163+ population recapitulated the CD1C_B/DC3 signature. (D) Proliferation of allogeneic CD4+ and CD8+ T cells 5 days after co-culture with CD14+ monocytes, pDCs, CD1C_A/DC2 DCs (CD1C+CD32B+), and CD1C_B DC3 (CD1C+CD163+). Left: Representative pseudocolor dot plot. Right: Bar graphs of composite data (n=3, mean ± SEM, *P<0.05, paired t-test).
