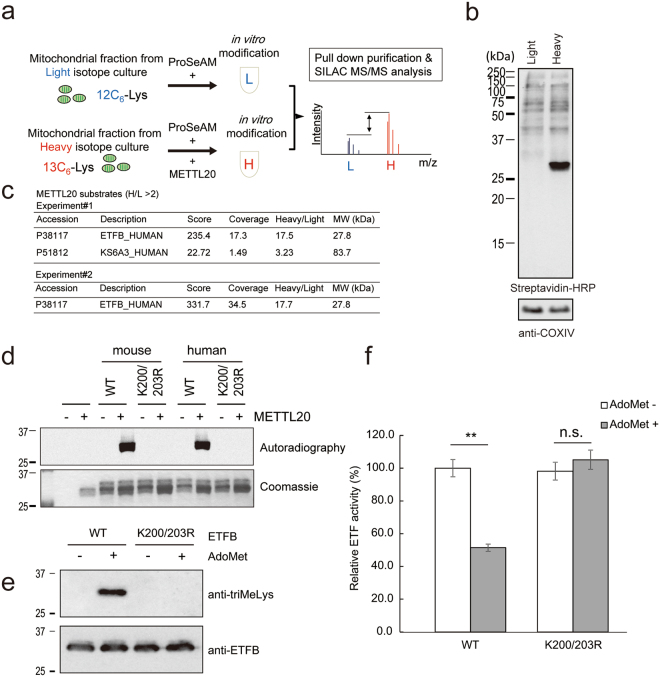
Figure 1.
ETFB methylation is conserved in human and mouse, which regulates the catalytic activity. (a) A schematic drawing of screening for METTL20 substrates. Cells were cultured in either light isotope labeled Lys containing medium (L) or heavy isotope labeled Lys containing medium (H). The mitochondrial lysates were reacted with ProSeAM with (H) or without (L) His-METTL20. After the in vitro modification, biotin tags were introduced to the modified residues via click reaction. Then the two samples were mixed together, and biotinylated proteins were pull-down with Streptavidin beads. After Lys-C digestion, peptides were analyzed by LC-MS/MS. (b) Confirmation of ProSeAM labeling. After the ProSeAM mediated alkylation and biotinylation via click reaction, small aliquot of the protein samples were analyzed by SDS-PAGE followed by western blotting with Streptavidin-HRP and anti-COXIV antibody as a loading control. (c) METTL20 substrates identified in the screening. Summary of two independent experiments were shown. Note that only ETFB was the protein identified in both cases. (d) Recombinant ETF complex (ETFA WT/ ETFB WT or K200/203R) and His-METTL20 were incubated with 14C-labeled AdoMet for 2 h at 30 °C. The autoradiography was detected with the image analyzer BAS-5000. (e) 4 µg of the ETF complex were mixed with 4 µg of METTL20 in the presence or absence of AdoMet and incubated at 30 °C for 3 h. After the reaction, they were mixed on ice with 8 µg of His-MCAD, 50 µM octanoyl-CoA and 70 µM DCPIP. Initial rate of the reduction of DCPIP was calculated from OD600 measured at 30 °C for every 30 sec for 10 min with a microplate reader (SpectraMax 190, Molecular Devices). Average of three independent experiments, n = 3 for each experiments, mean ± SEM. Student’s T-test **p < 0.01. (f) Small aliquot of the ETF used in A were separated with SDS-PAGE, and their methylation were detected with anti-triMeLys antibodies (top) and anti-ETFB antibodies (bottom, as loading control).