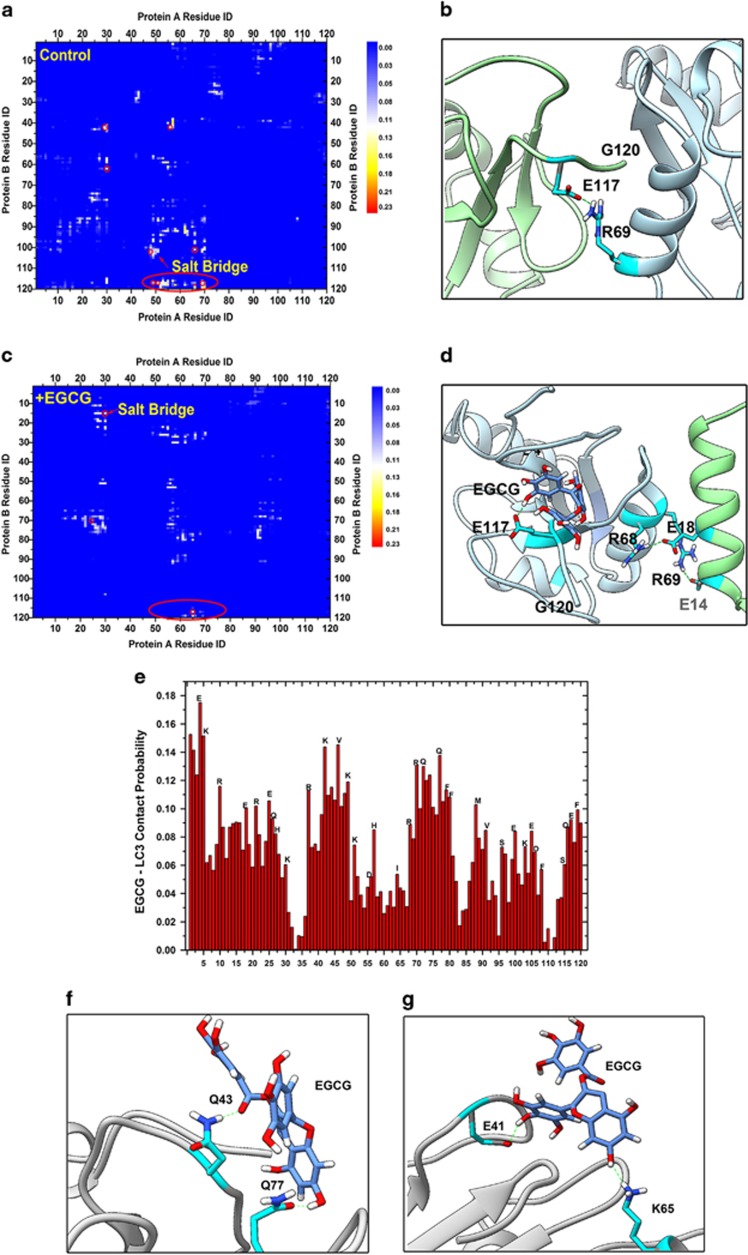
Figure 5.
The underlying molecular mechanisms for EGCG to inhibit the dimerization of LC3-I protein. The residual contact probability map of two LC3-I protein (a) is counted over all control systems, and (c) is counted over all the EGCG+ systems. The contact cutoff is defined as 0.5 nm. (b and d) The corresponding typical configurations highlighted by the red circle in (a) and (b), respectively. (e) The contact probability of EGCG with LC3-I protein (cutoff is 0.5 nm). The typical configurations for EGCG interacting with the polar residues (f), and the charged residues (g) through hydrogen bonds