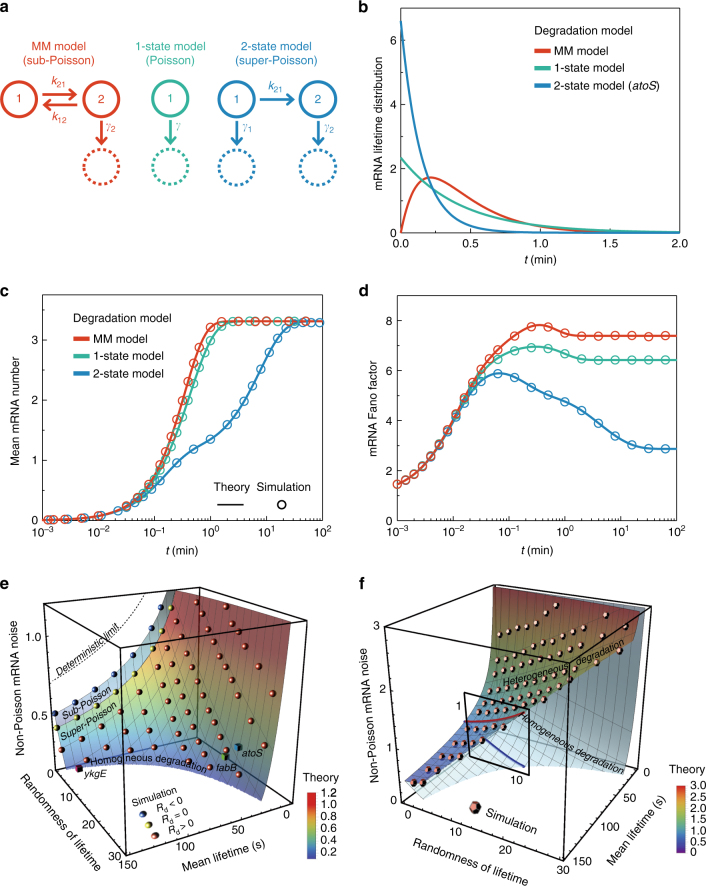
Fig. 5.
Quantitative prediction for mRNA noise dependence on mRNA lifetime fluctuation. a Models of the mRNA degradation process: (left) the Michaelis–Menten sub-Poisson process; (middle) one-state Poisson process; (right) two-state super-Poisson process. mRNA degradation starts from state 1. The three models represent renewal processes with different distributions of mRNA degradation time. Randomness, Rd, in mRNA degradation time or lifetime is defined by the relative variance of mRNA lifetime minus unity. For sub-Poisson mRNA degradation, Rd < 0; for a Poisson process, Rd = 0; and for a super-Poisson process, Rd > 0. b mRNA lifetime distributions. (red line) Michaelis–Menten (MM) sub-Poisson process; (green) 1-state Poisson process; (blue line) 2-state super-Poisson process (Supplementary Note 16). The 2-state super-Poisson model has been used to model the bi-exponential lifetime distribution of atoS, fabB, and ykgE mRNA63–65. The mean mRNA lifetime is set equal to 25.8 s, the mean lifetime of atoS mRNA. c, d Time dependence of the mean and Fano factor in the number of mRNA transcribed by a single gene. The mean and Fano factor of mRNA number are calculated from equation (M1-6) and Eq. (1) for Model III, optimized from our analysis in Fig. 2 for experimental data obtained from slowly growing E. coli36 (see Supplementary Table 1). The fraction of the active gene state is set to 1/2. e, f Dependence of the steady-state non-Poisson mRNA noise on the mean and randomness of the mRNA lifetime for two models: for the two-state super-Poisson mRNA degradation model without cell-to-cell heterogeneity (e) and for the 1-state Poisson mRNA degradation model, but with cell-to-cell heterogeneity in the rate (f). The theoretical prediction is made by the CFT in e, but by the generalized CFT, equation (M8-5) in f. (surface) Theoretical prediction. (spheres) Simulation results. (cubes) Prediction for atoS, fabB, and ykgE mRNA transcribed under IPTG inducible lac promoter in slowly growing E. coli shown in Fig. 2, given the 2-state super-Poisson model of mRNA degradation is valid66. Non-Poisson mRNA noise is measured by , or the mean mRNA number dependent component of non-Poisson mRNA noise produced by single-gene transcription (see Supplementary Note 6). Both models yield the same bi-exponential mRNA lifetime distribution. mRNA noise increases with the randomness in the mRNA lifetime originating from cell-to-cell heterogeneity in the mRNA degradation rate, but decreases with an increase in the randomness of mRNA lifetime caused by non-Poisson mRNA degradation dynamics (Supplementary Note 4 and Supplementary Movies 1 and 2)