Figure 3.
Rpap1 Knockdown Favors De-differentiation and Reprogramming
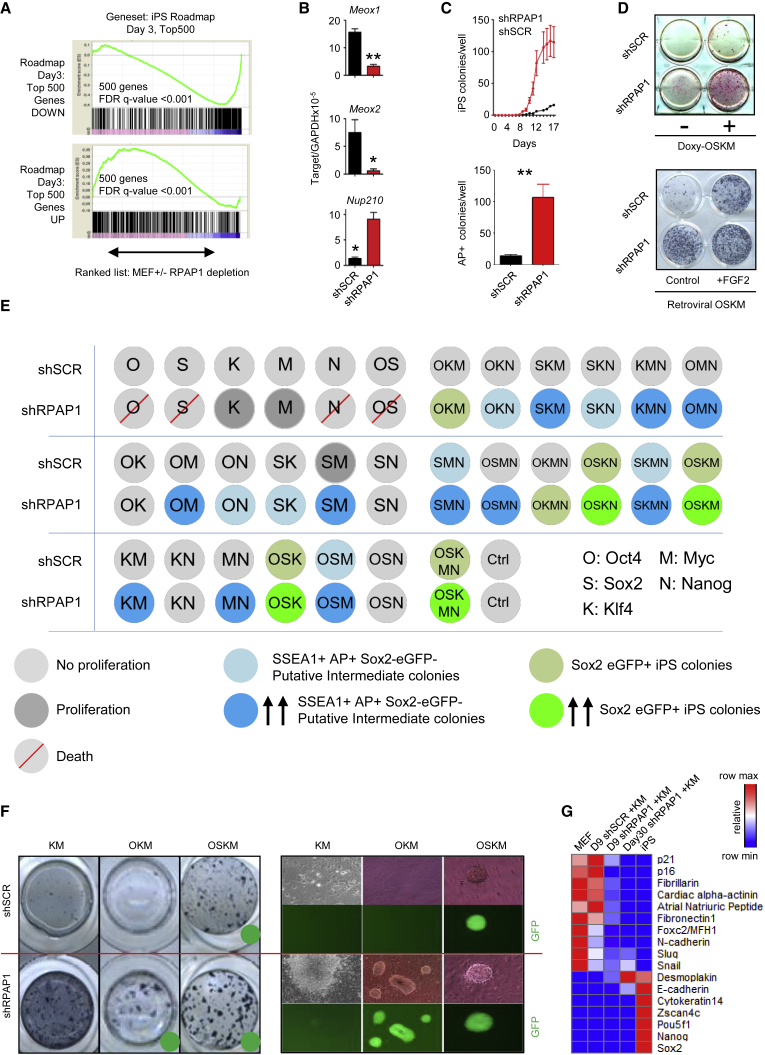
(A) Comparison of gene expression at day 3 after RPAP1 depletion in MEFs versus a published iPSC roadmap gene expression profile (Polo et al., 2012). Panels show GSEA comparison of the published top 500 genes up- or downregulated at day 3 of the iPSC roadmap versus a ranked list of the gene expression profile in the current study at day 3 after RPAP1 depletion in MEFs (x axis). See Supplemental Experimental Procedures for assessment of the iPSC roadmap data from parental MEFs verses Thy1-negative cells at day 3 of iPSC reprogramming. FDR q < 0.25 is significant. See also Figures S3A and S3B.
(B) qPCR measurement of selected genes at day 3 after RPAP1 depletion in MEFs. Downregulation of Meox1 and Meox2 and upregulation of Nup210 were reported to correlate with cell gene expression during the intermediate stages of iPSC reprogramming (Hansson et al., 2012, Polo et al., 2012). Mean ± SD; n = 3 independent MEF lines; ∗p < 0.05; ∗∗p < 0.01.
(C and D) MEF to iPSC reprogramming after RPAP1 depletion. Expression of the OSKM reprogramming factors was initiated at day 2 after lentiviral transduction of MEFs with non-targeting control (shSCR) or with RPAP1 targeting (shRPAP1) shRNAs. In (C), top panel, kinetics of iPSC colony appearance during doxycycline-induced reprogramming of i4F MEFs that express the four Yamanaka factors is shown (see Experimental Procedures). A profile representative of three independent i4F MEF lines is shown (mean ± SD; 3 technical replicates). In (C), bottom panel, quantification of iPSC colony yield at day 14 of doxycycline-induced 4F reprogramming is shown. Mean ± SD; n = 3 independent MEF lines; ∗∗p < 0.01. In (D), examples of alkaline phosphatase staining to indicate iPSC colonies formed at day 12 of i4F-MEF doxycycline-induced-OSKM iPSC reprogramming (top panel) or retroviral delivery of the OSKM factors (bottom panel) are shown. FGF2 was added to stimulate reprogramming efficiency.
(E) Summary of outcomes from 32 combinations of OSKMN Yamanaka transcription factors. Sox2-eGFP MEFs at day 2 after control or RPAP1 depletion received the indicated factors by retroviral delivery, followed by culture in standard iPSC reprogramming media. Progress of iPSC reprogramming was assessed by cell proliferation rate, morphology changes, colony formation, staining for alkaline phosphatase, SSEA1 expression, and Sox2-eGFP levels. Sox2-eGFP-positive cells forming typical iPSC colonies were scored as successfully reprogrammed iPSCs. Rapidly proliferating cells that initiated colony formation and that were positive for alkaline phosphatase and SSEA1 but negative for Sox2-eGFP were scored as putative intermediate stages of reprogramming.
(F) Examples of alkaline phosphatase staining to indicate formation rates of iPSC colonies and putative intermediate cell types at day 14 of MEF reprogramming with the indicated combinations of Yamanaka factors ± RPAP1 depletion. Green dot indicates those combinations that produced Sox2-eGFP-positive full reprogrammed iPSC colonies.
(G) qPCR measurement of mesenchymal, epithelial, and pluripotency marker mRNA expression levels. Data were converted to heatmap format to highlight the intermediate nature of marker expression displayed by the cells that were generated by shRPAP1+Klf4/Myc.
See also Figure S3.