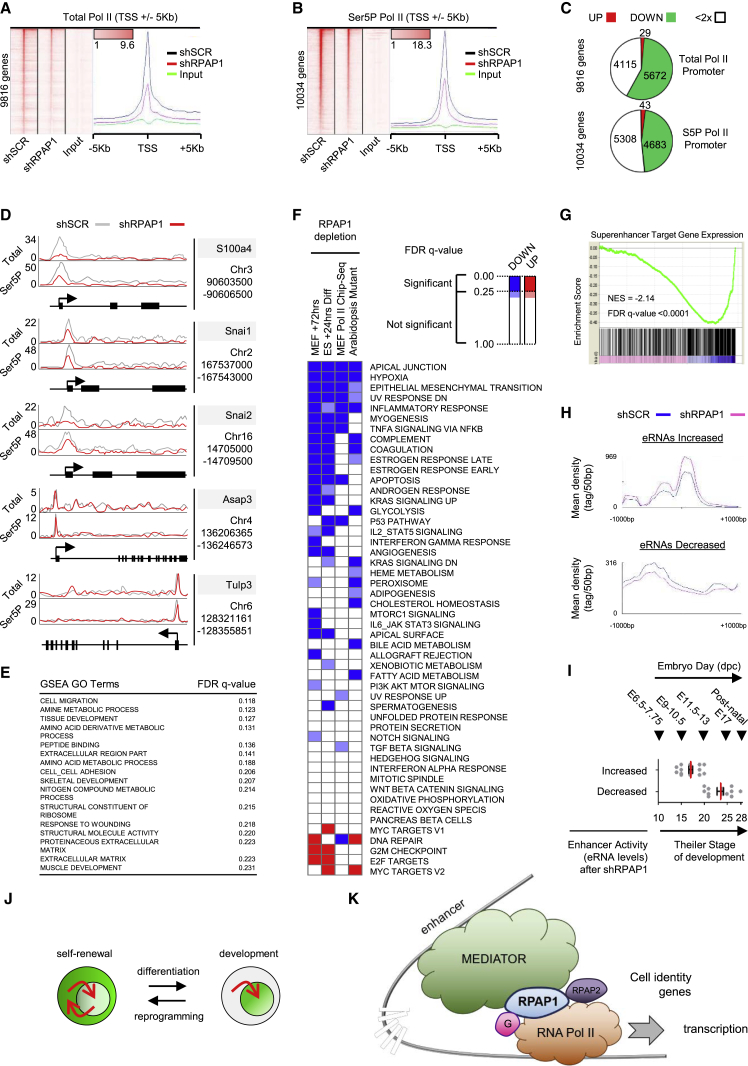
Figure 5.
RPAP1 Is Required for RNA Pol II Transcription in MEFs, Particularly on Developmental and Mesenchymal Genes
(A and B) ChIP-seq enrichment data after lentiviral transduction of MEFs with non-targeting control (shSCR) or with RPAP1 targeting (shRPAP1) shRNAs. Data are plotted as heatmaps of RNA Pol II total (A) or RNA Pol II Ser5P (B) occupancy around the transcriptional start site (TSS) region ± 5 Kb. Rows are sorted by decreasing RNA Pol II occupancy at the promoter (−100 to +300 bp) in the shSCR control. Color-scaled intensities are in units of reads per million mapped reads (rpm) (see Supplemental Experimental Procedures—ChIP-seq analysis).
(C) Proportional representation of ChIP data, classifying genes according to the changes in abundance of RNA Pol II total (upper panel) or Ser5P (lower panel) at the promoter (−100 to +300 bp) following RPAP1 depletion in MEFs for 3 days.
(D) Schematics of RNA Pol II total and Ser5P abundance on selected genes, showing examples of RNA Pol II depletion (S100a4, Snai1, and Snai2) or minimal effects (Asap3 and Tulp3).
(E) Table summarizing the most significantly up- or downregulated GO term gene sets identified by GSEA among the genes with >2×-fold decrease in RNA Pol II following RPAP1 depletion in MEFs (see also Tables S2 and S5). Gene sets with FDR q value < 0.25 were considered significant.
(F) Summary heatmap displaying the overlay of significant GSEA hallmark gene sets across 4 experiments (columns 1 and 2: see also Figure 2H). Column 1: GSEA on the ranked list of differential mRNA expression in MEFs at day 3 ± RPAP1 depletion is shown. Column 2: GSEA on the ranked list of differential mRNA expression in ESCs at +24 hr after inducing differentiation ± RPAP1 depletion is shown. Column 3: GSEA on the ranked list of differential RNA Pol II abundance at all promoters in MEFs at day 3 ± RPAP1 depletion is shown. Column 4: GSEA on the ranked list of differential gene expression in Arabidopsis thaliana plant tissues ± RPAP1 mutation is shown (Sanmartín et al., 2011), following conversion to the nearest mouse homolog based on protein sequence conservation (see Supplemental Experimental Procedures, conversion of plant to mouse homologs, and Table S2).
(G) GSEA to assess mRNA expression levels of MEF super-enhancer target genes (n = 661; defined by GREAT analysis as described; see Supplemental Experimental Procedures) within the transcriptome of primary MEFs at day 3 after Rpap1 knockdown. Compare with housekeeper gene expression in Figure S5G.
(H) Plots show the average eRNA levels within two groups of MEF super-enhancers regions: those which were increased (n = 63 enhancers; top panel) or decreased (n = 64 enhancers; bottom panel) in MEFs at day 3 after RPAP1 knockdown.
(I) GREAT analysis was used to identify target genes for each of the two super-enhancer groups identified in (H) (see Supplemental Experimental Procedures). These two sets of target genes (with increased eRNAs or with decreased eRNAs, respectively) were compared with gene groups associated to particular developmental Theiler stages (these gene groups are detailed in Figures S5H and S5I). The Theiler stage of those gene groups with high similarity to our target genes of interest is represented (gray dots) together with the average (red) and SEM (black).
(J and K) Model for RPAP1 function in the mechanism for triggering development.
(J) In self-renewing ESCs, RPAP1 (in green) is abundantly expressed and predominantly cytoplasmic. Upon differentiation, nuclear accumulation of RPAP1 permits increased transcriptional regulation and activation of developmental programs. Depletion of RPAP1 in differentiated cells results in loss of cell identity and enhanced susceptibility for reprogramming toward pluripotency.
(K) Taken together, our data suggest a model where RPAP1 exists in a complex with RNA Pol II and plays an essential role in the Mediator-RNA Pol II regulatory axis. Thus, loss of RPAP1 triggers a decrease in the association between Mediator and RNA Pol II (including the key regulators Gdown1 [G] and the Ser5P phosphatase RPAP2), preferentially affecting the ability of enhancers to activate Mediator target genes, which are known to include key markers and regulators of cell identity. In somatic cells, such as MEFs, this leads to de-differentiation, as expression of fibroblastic, mesenchymal, and developmental markers is erased.