Figure 2.
Performance Analysis of Organellar Maps Generated with TMT, LFQ, and SILAC Quantification Strategies
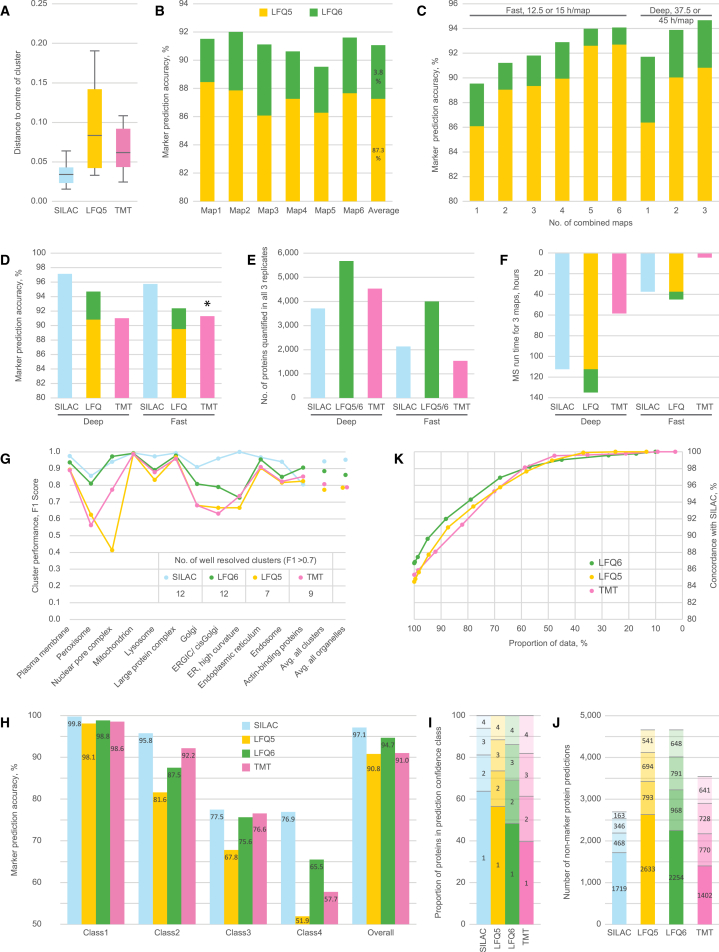
(A) To illustrate the relative precision of the different quantification methods applied in fractionation profiling, profile scatter within the 20S core proteasome (14 subunits, PSMA1–7, PSMB1–7, three independent measurements per protein) was analyzed (deep MS protocol). LFQ measurements are “noisier” than SILAC or TMT. Boxes indicate the interquartile range and whiskers 10th–90th percentile range.
(B) Organellar classification performance of six independent LFQ-based maps. Accuracy is the proportion of correctly classified organellar markers during supervised learning. Performance was assessed for six-fraction profiles (LFQ6, green) and for the same maps with the sixth data point removed (LFQ5, yellow).
(C) Combining several LFQ maps for organellar classification enhanced prediction accuracy. (Fast) maps shown in (B) were combined in the order of lowest to highest performance. Addition of each map improved performance. Maps 3, 4, and 6 were then chosen for further deep MS analysis and combined for classification.
(D) Marker prediction accuracy obtained with a combination of three replicate maps by quantification strategy and MS protocol. TMT fast maps included predictions for only 10 of 12 clusters (see also Figure S1G).
(E) Number of profiled proteins quantified in all three replicates.
(F) MS measurement requirements (hours) for the generation of three replicate maps.
(G–K) In-depth analysis of the predictions obtained with a combination of three replicate datasets, deep MS protocol (an equivalent analysis for predictions obtained with the fast MS protocol is shown in Figures S1B–S1E).
(G) Detailed performance profiles of maps made with SILAC, LFQ5/6, and TMT. Prediction performance was evaluated for each organellar cluster. F1 scores were calculated as the harmonic mean of recall (true positives / [true positives + false negatives]) and precision (true positives / (true positives + false positives]). High F1 scores (> 0.7) denote clusters with a high predictive value.
(H) Stratification of non-marker organellar predictions. Each assignment was made with a prediction confidence score. Four different SVM score cutoffs were defined, dividing the data into confidence classes. The prediction accuracy of marker proteins within each class served as a proxy for the prediction accuracy of non-marker proteins. Generally, the first two classes had high accuracies with all methods.
(I and J) Proportion (I) and absolute number (J) of non-marker predictions in each confidence class.
(K) Concordance analysis. The predictions of non-marker proteins, obtained with TMT, LFQ5, and LFQ6, were compared with the predictions obtained with SILAC. Concordance is the proportion of proteins with identical predictions. Restricting the comparison to proteins with a minimum confidence score in both compared maps reduces the overlapping dataset but increases concordance. In all cases, over 85% of the predictions show > 90% agreement.