Figure 3.
Assessment of Dynamic Organellar Maps with Different Quantification Strategies Using the Deep MS Protocol
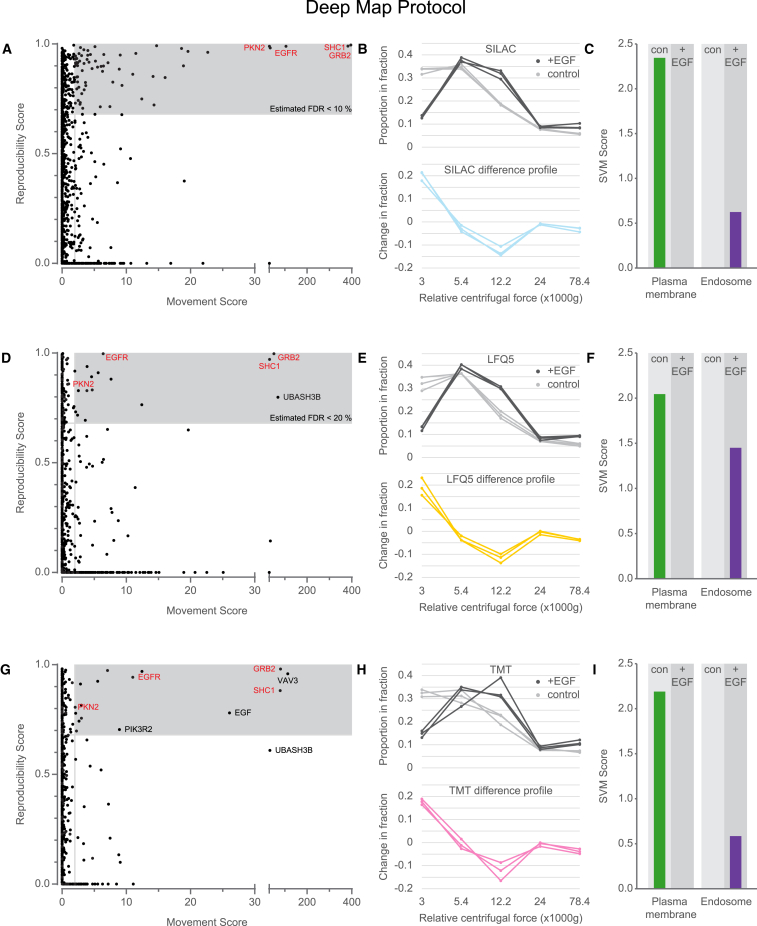
(A) Three replicate SILAC experiments of cells left untreated or stimulated with EGF for 20 min were analyzed in a dynamic organellar maps experiment. The resulting difference profiles were subjected to statistical analysis to identify moving proteins (see Experimental Procedures for details). The movement and reproducibility scores for each protein are shown in an MR scatterplot; significantly moving proteins have high scores in both dimensions. The shaded area contains proteins where the estimated false discovery rate (FDR) for translocation is < 10% based on a mock control experiment.
(B) Top: the proportion of EGFR in each fraction across the differential centrifugation gradient for three replicates in control cells (gray lines) or cells stimulated with EGF (black lines). Bottom: the difference in protein pelleting in the fractions in untreated compared with EGF-treated cells for three replicates.
(C) Proteins in the shaded area of (A) were removed from the marker set, and all remaining proteins were subjected to organelle classification using SVM-based machine learning. The prediction scores for the plasma membrane and endosome are shown before and after treatment with EGF, correctly capturing the change in localization of the EGF receptor.
(D–F) The same as (A)–(C), respectively, but for LFQ-based (deep) experiments. Note that the shaded area corresponds to a translocation FDR of < 20%.
(G–I) Also the same as (A)–(C), respectively, but using data from the TMT-based (deep) experiments. Note that the shaded area is not FDR-controlled but uses cutoffs determined for the SILAC and LFQ experiments.
See also Figures S2 and S3 and Table S2.