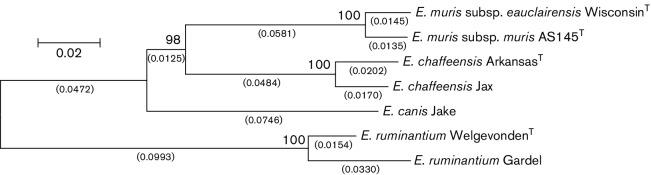
Fig. 1.
The evolutionary history was inferred using the neighbor-joining method [34]. The optimal tree with the sum of branch lengths=0.45356965 is shown. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown above the branches [35]. The tree is drawn to scale with branch lengths, shown under the branches (in parentheses), in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method [36] and are in the units of the number of base substitutions per site. The analysis involved seven nucleotide sequences and all positions with less than 95 % site coverage were eliminated. There were a total of 3781 positions in the final dataset. Evolutionary analyses were conducted in mega5 [37]. The following GenBank files were used in this analysis (E. chaffeensis ArkansasT genome NC_007799, E. chaffeensis Jax genome NZ_CP007475.1, E. canis Jake genome NC_007354, E. ruminantium WelgevondenT genome NC_005295.2, and E. ruminantium Gardel genome NC_006831.1).