Figure 6. Corrected rpoC1 and rps2 gene models.
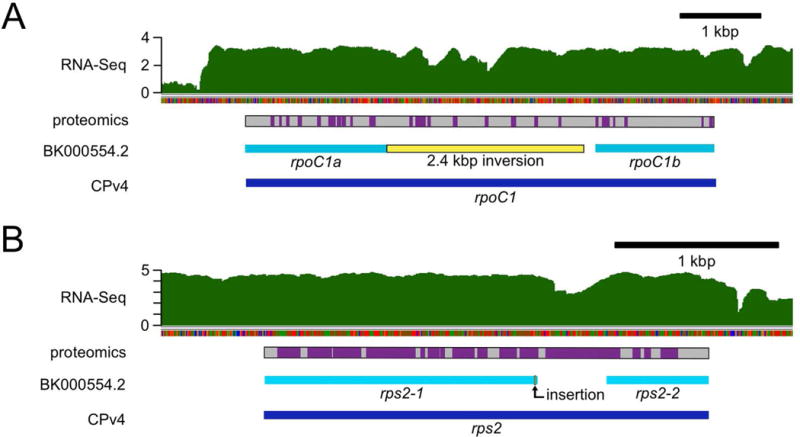
In the course of this work, we identified many errors in the genetic sequence of the previously available chloroplast genomes, such as GenBank BK000554.2. Some of these errors likely resulted in mis-annotated chloroplast genes. Here, RNA-Seq coverage and proteomics data were used to validate the improved CPv4 gene models. (A) In BK000554.2, the gene encoding the β′ subunit of the plastid-encoded RNA polymerase, rpoC1, was annotated as two separate genes: rpoC1a and rpoC1b (cyan bars). In contrast, CPv4 is annotated with a single ORF (dark blue bar) encoding a 1932 aa protein that spans both of the previous models. Relative to CPv4, there is a 2.4 kbp inversion (yellow bar) in BK000554.2 in the region between rpoC1a and rpoC1b. RNA-Seq coverage from a representative sample is presented on a log10 scale in green. A gray bar indicates the protein sequence encoded by CPv4 rpoC1, with purple boxes to indicate peptides that were identified by mass spectrometry. (B) Similarly, the gene encoding the S2 ribosomal protein was annotated as two genes in BK000554.2: rps2-1 and rps2-2 (cyan bars). A single nucleotide insertion (bent arrow) in that assembly causes a frame shift that leads to a premature stop codon. The CPv4 rps2 ORF (dark blue bar) spans both genes and encodes a 910 aa protein.
