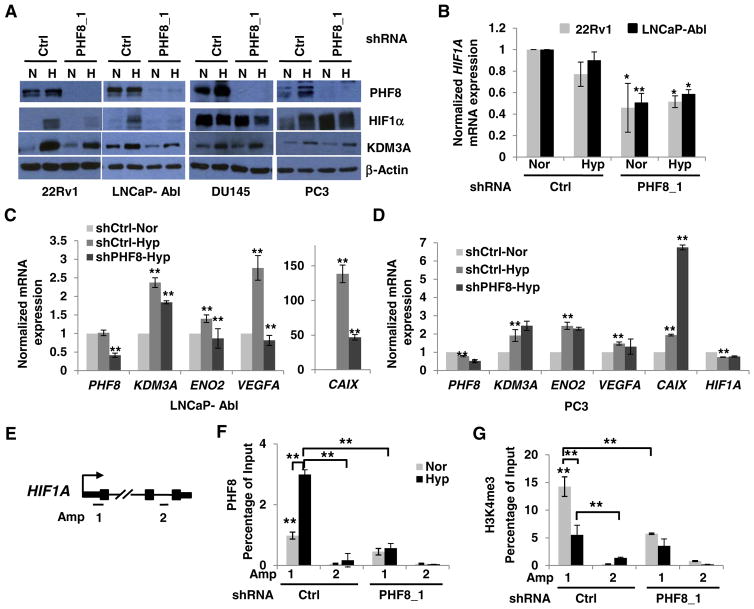
Fig. 6.
PHF8 regulates HIF1α and hypoxia-inducible genes in CRPC cells positive for full-length AR. A. Doxycycline-inducible control (Ctrl) or PHF8-targeted shRNAs were stably expressed in the indicated cell lines. The cells were induced by doxycycline for 72 h and cultured under normoxia (N or Nor) or hypoxia (H or Hyp; 1% O2) for 24 h. The indicated proteins were quantified by western blotting. B. HIF1A mRNA was assessed by RT-PCR in 22Rv1 and LNCaP-Abl cells. PHF8 RNAi cells were compared to controls undergoing the same treatment. C and D. The mRNA levels of the indicated genes were quantified by RT-PCR. Comparisons: between normoxia and hypoxia in the control RNAi; between PHF8 RNAi and the control RNAi cells under hypoxia. The S.D. was obtained from at least three independent experiments. Significance was calculated using the Student’s t-test. *: p < 0.05; **: p < 0.01. E. Schematic illustration of PCR amplicons to probe the Transcription Start Site (Amp 1) or non-specific control loci (Amp 2) of HIF1A gene. F and G. LNCaP-Abl cells stably expressing doxycycline inducible Ctrl or PHF8 shRNAs were cultured in normoxia (Nor) or 1% O2 hypoxia (Hyp) for 12 h. The ChIP-PCR data are interpreted as percent of 1% input DNA and the comparisons are as described in Fig. 3. The S.D. was obtained from at least three independent experiments. Two way ANOVA for multiple comparisons and Student t-test for paired comparison (s) were used to calculate the significance. *: p < 0.05; **: p < 0.01.