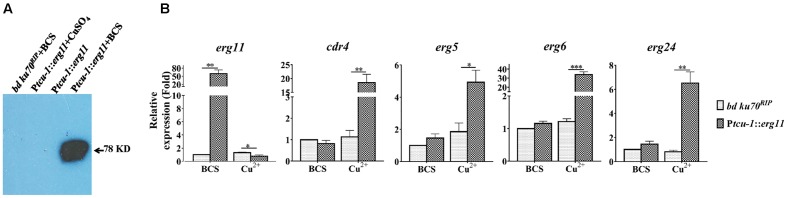
FIGURE 3.
Transcriptional induction of cdr4 and erg genes following erg11 inactivation. (A) Detection of ERG11 (sterol 14α-demethylase) by Western Blotting in the Ptcu-1::erg11 strain grown in Vogel’s medium supplemented with Cu2+ or BCS using anti-myc antibodies. The parental strain (bd Ku70RIP) grown in Vogel’s medium supplemented with BCS and the Ptcu-1::erg11 strain grown in normal Vogel’s medium was used as controls. (B) After grown in medium containing BCS for 13.5 h, mycelium were then shifted to indicated conditions to grow for additional 22 h. The transcript levels of selected genes were examined in the N. crassa bd Ku70RIP strain and Ptcu-1::erg11 strain treated with 50 μM BCS and 100 μM CuSO4 for 22 h. Transcript levels of cdr4 (NCU05591, encoding azole efflux pump CDR4), erg5 (NCU05278, encoding C-22 sterol desaturase), erg6 (NCU03006, encoding sterol C-24 methyl transferase), erg11 (NCU02624, encoding sterol 14α-demethylase) and erg24 (NCU08762, encoding C-14 sterol reductase) were measured by quantitative real-time polymerase chain reaction (qRT-PCR), and the expression was calculated by 2-ΔΔCt method and normalized to β-tubulin. The results presented here are means of three biological replicates, and the significant levels between the two strains in each treatment were calculated by t-test and marked as ∗(p < 0.05), ∗∗(p < 0.01) or ∗∗∗(p < 0.001).