Abstract
Genetic diversity is what selection acts on, thus shaping the adaptive potential of populations. We studied micro-evolutionary patterns of the key planktonic diatom Pseudo-nitzschia multistriata at a long-term sampling site over 2 consecutive years by genotyping isolates with 22 microsatellite markers. We show that both sex and vegetative growth interplay in shaping intraspecific diversity. We document a brief but massive demographic and clonal expansion driven by strains of the same mating type. The analysis of an extended data set (6 years) indicates that the genetic fingerprint of P. multistriata changed over time with a nonlinear pattern, with intermittent periods of weak and strong diversification related to the temporary predominance of clonal expansions over sexual recombination. These dynamics, rarely documented for phytoplankton, contribute to the understanding of bloom formation and of the mechanisms that drive microevolution in diatoms.
Introduction
Phytoplankton drive global biogeochemical cycles and regulate carbon fluxes in aquatic food-webs (Behrenfeld et al., 2005; D’Alelio et al., 2016). Eukaryotic microalgae reproduce asexually via mitosis and the sudden rise of cell division rates produces significant increases in the density of the population, called 'blooms' (Smayda, 1997; Wyatt, 2012).
Beside asexual reproduction, many unicellular eukaryotes include a sexual phase in their life cycle (Speijer et al., 2015), which has important ecological and evolutionary implications (von Dassow and Montresor, 2010). Through recombination, sexual reproduction introduces new genetic variants in a population on which selection can act, thus allowing evolutionary adaptations and shaping the population’s phenotypic profile. On the other hand, asexual reproduction allows the maintenance in the population of adapted genotypes in stable environmental conditions. However, genetic variation can arise in asexually reproducing species by means of mitotic mutations that can significantly contribute to the genetic standing variation in populations (Tibayrenc and Ayala, 2012).
Eukaryotic phytoplankton display a variety of life cycles, mating systems and ploidy levels; however, the role of these multifaceted life history architectures have in shaping genetic diversity is still poorly known (Rengefors et al., 2017). Theoretical models predict that even in predominantly asexual species, few recombination events per generation could be sufficient to give a population the same pattern of allelic variation as found in fully sexually reproducing organisms (Bengtsson, 2003). The rare occurrence of recombination has been invoked as the primary cause of the apparent dominance of clonal evolution, or clonality, in unicellular organisms, from bacteria to fungi and protozoans (Predominantly Clonal Evolution, PCE model (Tibayrenc and Ayala, 2012)). However, the PCE model mainly stems from observations carried out on non-free-living and pathogenic organisms. An alternative model of microevolution is the 'semi-clonal' or 'epidemic' model, in which recombination regularly occurs over long temporal scales but episodic clonal expansions can also occur (Smith et al., 1993; Maiden, 2006). The latter are short-term periods of both fast population increase and remarkable genetic divergence only driven by mutations. The population genetic structure of free-living cyanobacteria (D’Alelio et al., 2013), parasitic protozoa (Weedall and Hall, 2015) and fungi (MacLeod et al., 2000) have been interpreted in the frame of this latter model.
We assessed the relative impact of clonality (genetic divergence) and sexual recombination (genetic re-shuffling) on the population’s genetic structure of the marine planktonic diatom Pseudo-nitzschia multistriata, a marine planktonic species recurrently blooming at the Long-Term Ecological Research station MareChiara (LTER-MC) in the Gulf of Naples (Ribera d’Alcalà et al., 2004; D’Alelio et al., 2010; Tesson et al., 2014). Life history studies have revealed that P. multistriata has an heterothallic mating system: sexual reproduction can occur only when strains of opposite mating types (Mt+ and Mt−) are in contact (D’Alelio et al., 2009b). Occurrence of sex has been regularly observed in this species (D’Alelio et al., 2010) and intra-population recombination has been documented by ribosomal (D’Alelio et al., 2009a) and microsatellite (Tesson et al., 2013, 2014) markers. This species is also thought to be originated via hybrid speciation, that is, sex between congeneric species (D’Alelio and Ruggiero, 2015). Moreover, P. multistriata, as other species in this genus, produces the neurotoxin domoic acid, which represents a threat to human health and wildlife and a detriment to coastal aquaculture activities (McCabe et al., 2016; Berdalet et al., 2016).
We genotyped with 22 polymorphic microsatellites 519 strains of P. multistriata collected on a weekly basis in the Gulf of Naples (GoN, Mediterranean Sea) and in a few adjacent locations before and during the blooming seasons of 2 consecutive years (2013–2014). An unprecedented, clonal expansion involving mainly strains of the same mating type was recorded in the course of the summer-autumn bloom of 2013. Clear signatures of sexual recombination were recorded in the population collected in the pre- and post-bloom periods of 2013 and in the whole 2014. We also combined the data produced in this study (2013–2014) with a data set produced over 4 years (2008–2011) in a previous study (Tesson et al., 2014). This analysis allowed depicting a multiannual series of the genetic fingerprint of this diatom and gaining insights on its micro-evolutionary pathways.
Materials and methods
Strain isolation and cultivation
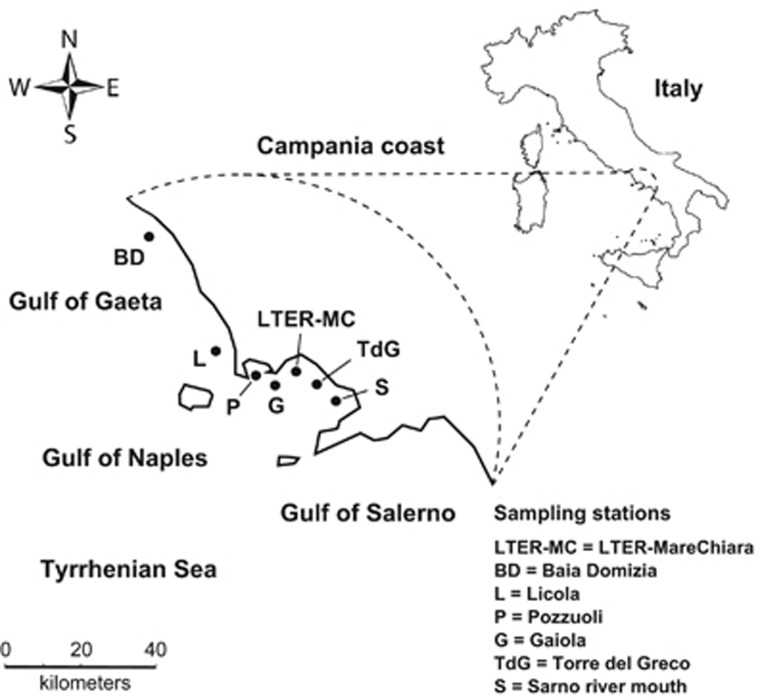
Single cells or short clonal colonies of P. multistriata were isolated from net samples, collected in the surface layer at the LTER-MC in 2013 and 2014 (LTER-MC, 40°48.5’ N, 14°15’ E, Gulf of Naples, Italy) and from seven locations along the coast between June and October 2013 and between July and October 2014 (Figure 1, Table 1). The cultures were grown in f/2 culture medium (Guillard, 1975) at a temperature of 18 °C, an irradiance of 50 μmol photons m−2 s−1 and a photoperiod 12:12 h L:D. For DNA analysis, within 10 days from strains' isolation, subsamples of cultures were harvested in late exponential phase by centrifugation and then processed as in Tesson et al. (2011).
Figure 1.
Map of the Campania coast (Tyrrhenian Sea, Mediterranean Sea) with the stations sampled in this study.
Table 1. Summary statistics for the samples of Pseudo-nitzschia multistriata collected in 2013–2014 and genotyped with 22 microsatellite markers.
| Sampling | N | MLG | RMLG | Na | Ho | Hnb | fIS | ISA |
|---|---|---|---|---|---|---|---|---|
| 5 February 2013 (MC) | 4 | 4 | 1.00 | 62 | 0.44 | 0.54 | 0.20 | −0.32 |
| 7 June 2013 (L) | 3 | 3 | 1.00 | 62 | 0.63 | 0.64 | 0.02 | 0.00 |
| 7 June 2013 (BD) | 7 | 5 | 0.67 | 59 | 0.52 | 0.49 | −0.08 | 0.22** |
| 16 July 2013 (MC) | 15 | 13 | 0.86 | 72 | 0.51 | 0.49 | −0.04 | 0.06** |
| 17 July 2013 (TdG) | 18 | 17 | 0.94 | 89 | 0.52 | 0.54 | 0.03 | 0.01* |
| 17 July 2013 (S) | 22 | 19 | 0.86 | 84 | 0.44 | 0.46 | 0.04 | 0.03** |
| 6 August 2013 (MC) | 4 | 4 | 1.00 | 51 | 0.54 | 0.50 | −0.12 | 0.00 |
| Pre-Bloom 2013 | 73 | 67 | 0.92 | 147 | 0.49 | 0.52 | 0.05 | 0.03** |
| 20 August 2013 (MC) | 47 | 47 | 1.00 | 129 | 0.55 | 0.50 | −0.10*** | 0.13** |
| 27 August 2013 (MC) | 11 | 10 | 0.90 | 47 | 0.57 | 0.35 | −0.66*** | 0.04** |
| 3 September 2013 (MC) | 37 | 32 | 0.86 | 93 | 0.57 | 0.46 | −0.25*** | 0.25** |
| 10 September 2013 (MC) | 10 | 9 | 0.89 | 54 | 0.63 | 0.41 | −0.57*** | 0.36** |
| 18 September 2013 (MC) | 28 | 27 | 0.96 | 83 | 0.54 | 0.41 | −0.33*** | 0.11** |
| Summer Bloom 2013 | 133 | 118 | 0.89 | 173 | 0.56 | 0.46 | −0.21*** | 0.16* |
| 24 September 2013 (MC) | 40 | 11 | 0.26 | 64 | 0.58 | 0.43 | −0.38*** | 0.31** |
| 24 September 2013 (P) | 11 | 3 | 0.20 | 36 | 0.55 | 0.36 | −0.80*** | 0.01 |
| 2 October 2013 (MC) | 61 | 7 | 0.10 | 45 | 0.64 | 0.39 | −0.74*** | 0.09** |
| 4 October 2013 (P) | 8 | 3 | 0.29 | 38 | 0.59 | 0.36 | −0.80*** | 0.02 |
| 4 October 2013 (G) | 7 | 5 | 0.67 | 44 | 0.50 | 0.39 | −0.34*** | 0.58** |
| 8 October 2013 (MC) | 38 | 10 | 0.24 | 60 | 0.59 | 0.41 | −0.46*** | 0.39** |
| 15 October 2013 (MC) | 28 | 4 | 0.11 | 50 | 0.48 | 0.43 | −0.14 | 0.18** |
| 22 October 2013 (MC) | 33 | 3 | 0.06 | 42 | 0.55 | 0.42 | −0.40*** | 0.60** |
| 28 October 2013 (MC) | 27 | 7 | 0.23 | 55 | 0.60 | 0.39 | −0.60*** | 0.14** |
| 7 November 2013 (MC) | 26 | 9 | 0.32 | 38 | 0.55 | 0.32 | −0.81*** | −0.01 |
| Autumn Bloom 2013 | 279 | 52 | 0.18 | 102 | 0.57 | 0.41 | −0.41*** | 0.22* |
| 1 July 2014 (MC) | 2 | 2 | 1.00 | 51 | 0.50 | 0.64 | 0.29 | na |
| 14 July 2014 (MC) | 3 | 3 | 1.00 | 61 | 0.50 | 0.56 | 0.13 | −0.03 |
| 2 September 2014 (MC) | 3 | 3 | 1.00 | 56 | 0.44 | 0.57 | 0.28 | 0.08 |
| 9 September 2014 (MC) | 13 | 12 | 0.92 | 95 | 0.61 | 0.64 | 0.04 | 0.04** |
| 7 October 2014 (MC) | 8 | 8 | 1.00 | 91 | 0.64 | 0.65 | 0.01 | 0.04** |
| 14 October 2014 (MC) | 3 | 3 | 1.00 | 62 | 0.58 | 0.63 | 0.12 | −0.01 |
| 21 October 2014 (MC) | 2 | 2 | 1.00 | 53 | 0.52 | 0.64 | 0.25 | na |
| 2014 | 34 | 34 | 1.00 | 139 | 0.58 | 0.65 | 0.10 | 0.02* |
For each sample are given: sampling date and location (MC, Long-Term Ecological Research station MareChiara; BD, Baia Domizia; G, Gaiola; L, Licola; P, Pozzuoli; S, River Sarno mouth; TdG, Torre del Greco; for geographic locations of sampling sites, see Figure 1), number of genotyped strains (N), number of multilocus genotypes (MLG), genotypic richness based on MLG (R=(MLG−1)/(N−1)), number of alleles over all loci (Na), observed (Ho) and non-biased expected (Hnb) heterozygosity, fixation index (fis) and association index over all loci (ISA). Significance values for FIS and ISA are as follows: *P<0.05; **P<0.01; ***P<0.001. na= not applicable, Ho, Hnb, fis and ISA were calculated excluding all but one replicate of each MLG per sample. Sum or average values for the different periods are marked in bold.
Selection and development of microsatellite loci
The strains isolated in the present study (2013–2014) were analyzed with 22 microsatellite markers, seven designed by Tesson et al. (2011) and 15 new microsatellite loci identified in silico on the P. multistriata genome assembly (Basu et al., 2017). All possible di- and tri-nucleotidic sequence combinations (AC, AAC, AG, AAG, AT, AAT, CG, ACC, ACG, ACT, AGC, AGG, ATC, CCG, excluding frame-shifts and reciprocal reverse-complements), were searched in the genome using the Blast application (Altschul et al., 1990). As microsatellites are repetitive elements, it was necessary to include low complexity regions in the blast analysis. Primers were designed using the web-based software Primer3 Plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi). The size of the resulting fragments was chosen to maximize the possibility to set-up multiplex PCR reactions. Forward primers were labeled with four different fluorochromes (PET, NED, 6FAM and VIC, Life Technologies, Carlsbad, CA, USA). The 22 microsatellite loci were amplified in multiplex reactions. Multiplex composition is shown in Supplementary Table 1; up to six loci were amplified in the same reaction.
Amplification reactions were performed using the Type-it Microsatellite PCR Kit (Qiagen Ltd., Venlo, The Netherlands), according to manufacturer instructions. Microsatellite reactions were prepared in automation with a robotic station BIOMEK FX (Beckman Coulter, Fullerton, CA, USA) and analyzed on an Automated Capillary Electrophoresis Sequencer 3730 DNA Analyzer (Life Technologies). Size of alleles was determined using the software PEAK SCANNER v1.0 (Applied Biosystems, Foster City, CA, USA). The species specificity of the primers was tested in silico. The sequence of the flanking regions of each locus was blasted against other three available diatom genomes (Fragilariopsis cylindrus, Phaeodactylum tricornutum and Thalassiosira pseudonana) and against the GenBank database (NCBI BLAST version 2.2.9, (Altschul et al., 1990)).
Microsatellite data analysis
The data set analyzed for microsatellite analyses was built allowing a maximum of 10% of missing loci per individual multilocus genotype (MLG). This threshold was obtained based on the methodological approach by Pemberton et al. (2013). The presence of null alleles under the assumption of Hardy–Weinberg equilibrium was verified using the software Microchecker (version 2.2.3) (Van Oosterhout et al., 2004). The frequency of null alleles was estimated using the Brookfield-1 estimator implemented in the software. The presence of stutter bands and of large allele dropouts was also investigated. MLGs were screened for somatic mutations using the software Genclone v2.0 (Arnaud-Haond and Belkhir, 2007), through the identification of pairwise differences.
Genotypic richness was calculated as:
where MLG is the number of genotypes and N is the total number of strains in the sample. Total expected heterozygosity (Ht) for each locus were obtained in Genalex v.6.5 (Peakall and Smouse, 2006). Observed and unbiased expected heterozygosity (Ho and Hnb, respectively) per sample and the fixation index (fis) were calculated using Genetix (Version 4.0, http://kimura.univ-montp2.fr/genetix/). The significance of fis was estimated using a permutational approach: 1000 permutations of the alleles within samples were performed. Within each sample, all but one of the replicated MLGs were eliminated, to avoid artifactual deviations from Hardy–Weinberg equilibrium. Hardy–Weinberg equilibrium within samples was tested using Genepop v. 4.2 (Raymond and Rousset, 1995) (see also http://genepop.curtin.edu.au/).
The index ISA (standardized index of association), that estimates the overall linkage disequilibrium per sampling date, was calculated using LIAN 3.7 ((Haubold and Hudson, 2000) see also http://guanine.evolbio.mpg.de/cgi-bin/lian/lian.cgi.pl/query). The software LIAN tests for independence of allele assortment, computing the number of loci at which two MLGs differ and then compares the observed variance of the mismatch values (Vd) with respect to the variance expected under linkage equilibrium (Ve). ISA is calculated as:
where n is the number of loci.
A restricted data set without missing data was built to calculate (i) allelic distance between genotypes, (ii) the probability of occurrence of a given MLG (Pgen(fis)) and (iii) the probability the genotype to be present >1 to n times or more as the result of distinct sexual events (Psex(fis)) (the latter probabilities were calculated taking into account the Hardy–Weinberg equilibrium departures). The final allelic distance matrix included 120 MLGs. This data set was analyzed using a network approach (See Supplementary Methods).
A principal coordinate analysis was performed in Genalex v.6.5 (Peakall and Smouse, 2006), using as input a genotypic–distance matrix produced for each sample. To test the population genetic structure on a longer temporal scale, we analyzed the genetic data obtained in the present study and those obtained by Tesson et al. (2014), a study on P. multistriata populations genetic structure in the GoN for the years 2008–2011. Different methods of fragment analysis were used in the two studies and therefore a standardization procedure was applied by re-amplifying a selection of strains with the current fragment analysis methods. The standardization resulted in reliable comparisons between allele reads for five loci common to the two studies, namely: PNm1, PNm2, PNm3, PNm7, PNm16.
Methods for the experiments aimed at testing (i) the presence of parasites, (ii) differences in growth rates and (iii) competition between MLG#86 and other genotypes are in Supplementary Methods.
Results and discussion
Clonality versus recombination at short time scales
P. multistriata was first detected in the GoN in 1994 and, since then, it is regularly recorded during summer and autumn (D’Alelio et al., 2010; Ribera d’Alcalà et al., 2004; Tesson et al., 2014). This was also the time at which the species was recorded during our study in the years 2013 and 2014 (Figure 2b).
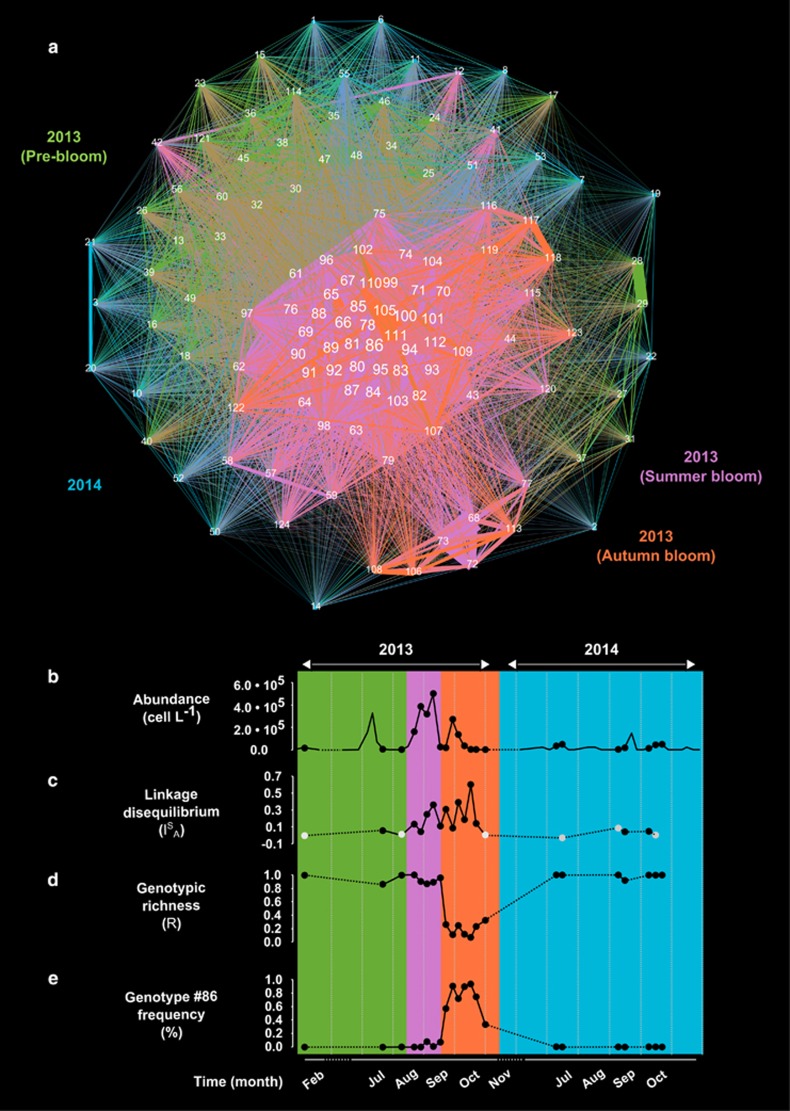
Figure 2.
Population genetic structure of Pseudo-nitzschia multistriata in the Gulf of Naples. The different colors mark samples collected in different periods. (a) Similarity network: nodes represent multilocus genotypes (MLGs); the width of lines connecting nodes is proportional to allelic similarity between nodes; colors identify the period of peak abundance of each MLGs (green=2013 pre-bloom; pink=2013 summer bloom; orange=2013 autumn bloom; blue=2014); the size of the label of each node is proportional to the level of connection with neighboring nodes (weighted degree). (b) Trends of P. multistriata cell abundances at LTER-MC; sampling dates for population genetics are marked with a black circle. (c) Linkage disequilibrium (parameterized by the association index ISA); bright gray circles indicate not statistically significant values. (d) Genotypic richness (R). (e) Frequency of MLG #86.
A total of 443 strains, collected at LTER-MC on 22 sampling dates, and additional 76 strains collected at other seven localities along the coast (Table 1,Figure 1) were genotyped with 22 microsatellite markers. A total of 271 MLGs containing up to 10% of missing (that is, not amplified) loci per genotype were recorded (Table 1). The relatively low genotypic diversity was due to the overrepresentation of few MLGs (see below). After including only the MLGs where all loci were successfully amplified, a total of 120 MLGs was recovered.
The reciprocal relatedness of these MLGs was studied with a network approach in which MLGs were the nodes and the microsatellites-based allelic similarities among MLGs were the links (Figure 2a). The resulting network showed the presence of two distinct modules, that is, ensemble of nodes whose reciprocal similarity is higher than the similarity between them and the rest of the network. A first outer module was broadly scattered and included MLGs occurring mainly during the pre-bloom phase of 2013 (green links in Figure 2a) and in 2014 (blue links in Figure 2a). A second inner module included only MLGs occurring during the summer-autumn blooms of 2013 (orange and violet links in Figure 2a). MLGs in the latter module differed for less than 10 alleles (average allelic similarity: 0.11), whereas a lower average similarity between MLGs (0.04, corresponding to 26 alleles) was observed in the first module. The most reasonable explanation for the higher genotypic similarity between MLGs in the inner module is that during a period of fast vegetative replication, as a bloom is, mitotic mutations take place very quickly, as in a typical clonal expansion (Smith et al., 1993; Maiden, 2006). Remarkably, about one fourth of the overall diversity (10 out of 39 alleles of differences among all MLGs) was attributable to mutations emerged during summer and autumn blooms of 2013. A high microsatellite mutation rate was indeed already recorded in P. multistriata clonal strains grown in the laboratory over several months (Tesson et al., 2013). Within the inner network module, MLG #86 was the node with the highest degree of interconnection: this suggests that this genotype had a pivotal role in the clonal evolution of MLGs detected in summer and autumn 2013.
In Figures 2b–e, the abundance of P. multistriata at LTER-MC is represented together with linkage disequilibrium (parameterized by the index ISA). Linkage disequilibrium provides the degree of association among alleles of multiple loci, which are neutrally evolving and free from strong selective pressures (Tibayrenc and Ayala, 2012), such as microsatellite markers. In a panmictic sexual population, meiotic recombination results in the free exchange of alleles, which produces their random association (linkage equilibrium, ISA close to zero), whereas the rarity or absence of sex results in the dominance of clonal divergence over genetic recombination (linkage disequilibrium, 0<ISA<1) (Schurko et al., 2009b). Although linkage disequilibrium for the 2013 pre-bloom (green portion of the graph in Figure 2c) was negligible (ISA close to 0), it considerably increased during the summer and autumn blooms (ISA up to 0.6; Figure 2c). This indicates a progressive strengthening of the association between MLGs owing to the dominance of genetic mutations, corroborating the view that MLGs in the inner module of the network (Figure 2a) producing blooms during summer and autumn 2013 (Figure 2b) evolved predominantly clonally. Eventually, linkage disequilibrium dropped again in 2014, indicating the re-establishment of a frequent recombination state. The association index ISA for all the investigated period was calculated on single MLGs, that is, excluding replicates, so to rule out overrepresentation of some genotypes with respect to others. The presence of linkage disequilibrium in the P. multistriata population during summer and autumn 2013 was flanked by significantly negative fis values, indicating heterozygosity excess (Table 1), which denotes fast intra-population genetic divergence driven by mitotic mutations, as is the case of sudden clonal expansions (the 'Meselson effect' (Stoeckel and Masson, 2014)). Conversely, a balance between expected and observed heterozygosity, indicating the presence of widespread recombination, was recorded in the 2013 pre-bloom phase and in all samples collected in 2014 (Table 1).
The micro-evolutionary changes occurring during the 2013 blooms induced relevant modifications in P. multistriata population diversity, which was described in terms of genotypic richness (Figure 2d). At the beginning of the 2013 summer bloom (violet portion of Figure 2d), genotypic richness was close to 1. This parameter slightly decreased as soon as linkage disequilibrium increased, suggesting that the reduction in richness was mainly due to the rise in dominance of lineages evolved clonally. Afterwards, richness decreased sharply during the autumn bloom (orange portion of Figure 2d) and came back to maximum values in the course of 2014. The brisk fall in genotypic richness in autumn 2013 was due to the numerical dominance of MLG #86 (Figure 2e). This genotype first appeared on 3 September 2013, peaked in October, and it was no longer recorded in the subsequent year. The probability of occurrence (Pgen(fis)) and the probability that identical MLGs may have arisen by chance more than once from distinct sexual events (Psex(fis)) for the dominant genotype were, respectively, as low as 3.12 × 10−05 and 0, when up to 73 genotyped strains shared the same MLG (Supplementary Table 2). These parameters strongly indicate that all genotyped strains displaying MLG #86 (n=208) were actually part of a single clonal lineage, evolved in the course of a clonal expansion.
Knowledge of the life cycle of P. multistriata helps in shedding light on the micro-evolutionary processes in this planktonic diatom. Our model species has a heterothallic mating system and it is thus possible to attribute the mating type to clonal strains isolated from the sea (Supplementary Text). Although the sex ratio of strains isolated in 2014 was ~1:1 (n= 29), out of the 215 strains tested for mating type in 2013, 92.6% were Mt+, including MLG #86 and its closely related genotypes. This strongly unbalanced sex ratio further supports the fact that sex was not frequent in that period and that all genetic variants around MLG #86 (in the violet and orange portion of the network in Figure 2a) originated via mitotic mutations. In the light of the results illustrated above, we can summarize that: (i) the 2013 pre-bloom population was characterized by high standing genetic diversity under a condition of free-recombination; (ii) a ‘cloud’ of MLGs, reciprocally differing for a few alleles, rose to dominance and started diverging clonally in absence of sex during the 2013 summer bloom; (iii) MLG #86 evolved from this first clonal burst; (iv) in the course of the autumn bloom, MLG #86 expanded demographically and clonally, generating a second group of MLGs via mutations; (v) the ‘clonal cloud’ dissolved and the free-recombination state was re-established.
To the best of our knowledge, this is the first report of a dramatic event of clonal expansion for a marine planktonic protist. High genotypic diversity has generally been detected for aquatic microalgae (Rengefors et al., 2017), including diatoms sampled along a bloom such as Ditylum brightwellii (Rynearson and Armbrust, 2005), Skeletonema marinoi (Godhe et al., 2016) and P. multistriata (Tesson et al., 2014 and this study). Nonetheless, low diversity was observed in some cases: the dominance of asexual reproduction and clonal evolution were considered at the base of the relatively low genotypic richness (0.667) reported for the coccolithophore Emiliania huxleyi during a bloom in the North Sea (Krueger-Hadfield et al., 2014). Extremely low genotypic richness (down to ~0) was also reported for a benthic freshwater diatom collected in small pond (Vanormelingen et al., 2015), although the hypothesis of clonal expansion could not be investigated due to the limited number of samples.
To test for the presence of selective factors that may have determined the rise of clonal lineages during summer and autumn 2013, we tested for the presence of parasites that can represent selective forces in diatoms, where parasitic chytrids have been reported to induce population differentiation (Gsell et al., 2013). Yet, we could not find chytrids in P. multistriata cells collected before and during the 2013 bloom of P. multistriata. We also measured maximum growth rates in monoculture of strains belonging to the dominant MLG #86 and to different MLGs but no significant difference in their growth capability was detected (t-test, P>0.05) (Supplementary Table 3). We conducted a competition experiment based on mixed cultures started with equal proportions of one strain with the profile of MLG #86 and three strains with a different genetic profile (but all belonging to the inner violet-orange module in Figure 2a): at the end of the culturing period, all 26 strains sampled from the mixed cultures belonged to MLG #86. These results suggest that this genotype might have a superior fitness, but these preliminary data do not allow drawing hypotheses on the mechanisms at the base.
Population genetic structure
Population genetics studies of planktonic species sampled at fixed monitoring stations are challenging due to the intrinsic patchiness of the pelagic environment. For instance, patches of different populations can be sampled at different times and these patches can include distinct genotype assortments.
To rule out this potential bias, a spatial sampling was also carried out in summer 2013: strains were collected at other stations falling in a geographical range of about 50 km centered on LTER-MC (Figure 1; Table 1). The population genetic structure of P. multistriata in the GoN and nearby locations in the course of years 2013–2014 is shown by the principal coordinate analysis illustrated in Figure 3a. The principal coordinate analysis clearly showed that the genetic diversification gained by P. multistriata was mainly dependent on the sampling time. In fact samples collected outside the long-term station LTER-MC, represented by triangles in Figure 3a, clustered together with the LTER-MC samples collected in the same periods. This shows that a single population extending well beyond LTER-MC was sampled in the course of the present investigation (Figure 3a).
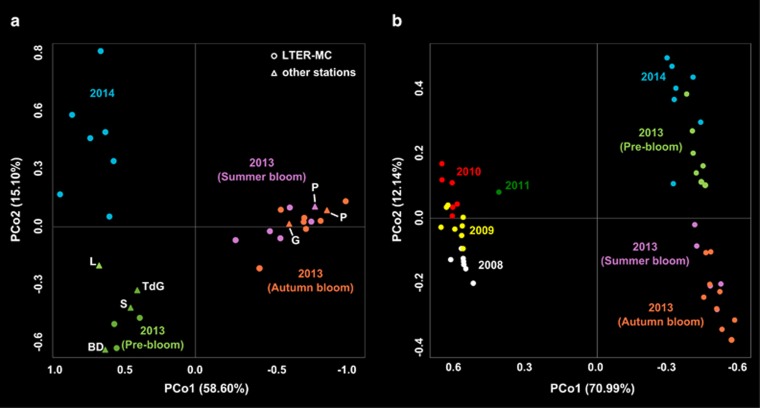
Figure 3.
Population differentiation of Pseudo-nitzschia multistriata (a) PCoA of population differentiation in time, based on 22 microsatellites, among samples collected at LTER-MC and other sites over 2013–2014; color coding as in Figure 2. (b) PCoA of population differentiation in time, based on five microsatellites, among samples collected at LTER-MC over the period 2008–2014; colors refer to different time periods. Percentages of variance are indicated in brackets at each PCo axes.
The brisk clonal expansion in the 2013 blooms of P. mulstriata determined a remarkable leap of the population fingerprint along the PCo1 axis, whereas the 2013 pre-bloom and the population collected in 2014 diverged along the PCo2 axis. A clear differentiation could be observed between samples from the pre-bloom period and samples from the blooming season (summer+autumn) and both were differentiated from 2014 samples. We could have hypothesized that the population differentiation occurred in 2013 was driven by environmental factors, such as temperature, salinity or nutrients, but this is not the case, since physical-chemical parameters had similar trends in 2013 and 2014 summer-autumn periods (Supplementary Figure 1).
A multiannual analysis of population genetic structure
In order to gain insights on the long-term patterns of the P. multistriata population in the Gulf of Naples, we merged the data set produced in the present study with the data set of Tesson et al. (2014), which included strains sampled at LTER-MC in 2008–2011 (Figure 3b). The population of P. multistriata was divided into two main groups in relation to PCo1: the first group included samples from 2008–2011, the second those from 2013–2014 (Figure 3b). In the course of 2013–2014, the diversification along the PCo2 was wider than in the preceding years. The population structure of P. multistriata thus changed over time with a nonlinear pattern and with the alternation between one period of weak (2008–2011) and one of strong diversification (2013–2014). Remarkably, the lowest values of genotypic richness (R=0.385) gathered in 2008 (Table 2) corresponded to a marked dominance of one mating type (Mt+) (Supplementary Text). Moreover, we also recorded an unbalanced ratio between expected and observed heterozygosity and relatively high linkage disequilibrium in 2008: these observations suggest deviation from a frequent recombination state likely analogous to that recorded in 2013. Our observations suggest that episodic demographic expansion of genotypes of the same mating type could be an important mechanism in clonal evolution, and thus population differentiation, in P. multistriata.
Table 2. Summary statistics for the samples of Pseudo-nitzschia multistriata collected in the present study (2013–2014) combined with the data collected by Tesson et al. (2014) in 2008–2011; all samples were genotyped with five microsatellite markers.
| Sampling year | N | MLG | RMLG | Na | Ho | Hnb | fIS | ISA |
|---|---|---|---|---|---|---|---|---|
| 2008 | 157 | 61 | 0.385 | 34 | 0.656 | 0.574 | −0.144*** | 0.037* |
| 2009 | 193 | 107 | 0.552 | 30 | 0.570 | 0.572 | 0.004 | 0.014* |
| 2010 | 162 | 124 | 0.764 | 35 | 0.648 | 0.592 | −0.095*** | 0.007 |
| 2011 | 13 | 12 | 0.917 | 18 | 0.617 | 0.523 | −0.190* | 0.002 |
| 2013 | 485 | 110 | 0.225 | 33 | 0.566 | 0.523 | −0.083*** | 0.012* |
| 2014 | 34 | 33 | 0.970 | 22 | 0.604 | 0.559 | −0.084 | 0.021 |
For each sample are given: sampling year, number of genotyped strains (N), number of multilocus genotypes (MLG), genotypic richness based on MLG (R=(MLG−1)/(N−1)), number of alleles over all loci (Na), observed (Ho) and non-biased expected (Hnb) heterozygosity, fixation index (fis) and association index over all loci ISA. Significance values for fis and ISA are as follows: *P<0.05; **P<0.01; ***P<0.001. na= not applicable, Ho, Hnb, fis and ISA were calculated excluding all but one replicate of each MLG per sample.
We also have to consider the demographic dynamics of diatom populations: the increases of abundance during the bloom seasons are followed by severe bottlenecks in which the species are not detected in the water column. Benthic resting stages that can act as ‘seed beds’ for the inoculum of subsequent blooms are apparently not produced by Pseudo-nitzschia species and it is thus reasonable to assume that rare cells remain in the water column between the blooms. The genetic fingerprint of the survivor population determines the genetic profile of the next bloom. Considering the rate at which populations diversify mitotically and the frequent recombination events, it is not surprising that the genetic make-up can considerably change from one year to another.
To date, few studies addressed the temporal scale of population genetic structure in planktonic protists. Interannual genetic differentiation was recorded in two species of the dinoflagellate genus Alexandrium and was attributed either to markedly fluctuating environmental selection or to the germination of distinct pools of benthic resting cysts (Erdner et al., 2011; Richlen et al., 2012; Dia et al., 2014). Genetic differentiation was observed in populations of the diatom Skeletonema marinoi sampled during the seasonal bloom in a Swedish Fjord (Saravanan and Godhe, 2010) and in the Baltic Sea (Godhe et al., 2016). In contrast to the previous studies, a surprisingly stable genetic structure was recorded in a population of S. marinoi by analyzing strains obtained from the germination of resting stages from core sediment samples dating back 100 years (Härnström et al., 2011). Intra-population gene flow, exerted by frequent sexual events in a relatively secluded environment, may explain this long-term genetic homogenization in S. marinoi (Godhe et al., 2014). Among diatom populations there is thus evidence for both short-term differentiation (Saravanan and Godhe, 2010; Vanormelingen et al., 2015; Godhe et al., 2016), as also reported in the present study and likely driven by clonality, and long-term homogenization (Härnström et al., 2011), driven by sexual recombination.
Diatom clonality and sex in relation with evolutionary models
Two main theoretical models of genetic diversification at species and population levels have been developed for unicellular organisms to reconcile the apparently antithetical roles of clonality and recombination in driving diversification: the 'predominantly clonal evolution (PCE)' (Tibayrenc and Ayala, 2012) and the 'semi-clonal’ or ‘epidemic' evolution model (Maiden, 2006; Smith et al., 1993). Within PCE, clonality in predominantly asexual or autogamic organisms is pursued by limited recombination among clonal lineages by means of intrinsic biological mechanisms (Tibayrenc and Ayala, 2012). For instance, genomic mechanisms regulating shifts between clonality and sexuality have been detected in the predominantly parthenogenetic genus Daphnia in form of a diallelic system (Schurko et al., 2009a). Conversely, in the epidemic model, recombination is free and clonal evolution occurs after selective processes (Smith et al., 1993; Maiden, 2006).
Concerning phytoplankton, frequently recombining populations of the freshwater cyanobacterial genus Plankthotrix showed epidemic clonality (that is, local expansions of clonal lineages) caused by selective pressure exerted by lakes circulation regimes over genes regulating buoyancy (D’Alelio and Gandolfi, 2012; D’Alelio et al., 2013). Epidemic evolution has been invoked also in unicellular eukaryotes, such as parasitic protozoa (Weedall and Hall, 2015) and fungi (MacLeod et al., 2000). Based on the results of the present study, the PCE model cannot be applied to fully explain the genetic dynamics of P. multistriata because sex and genetic recombination regularly occur, as shown by linkage equilibrium values and the balance between expected and observed heterozygosity. However, within this context of regular recombination, clonal expansion events can also occur, as shown by the dominance of MLG #86 and its cloud of closely related genotypes, and thus the semi-clonal or epidemic model better represents the population dynamics of this diatom. The dominance of a clonal lineage is usually explained by its superior fitness. The results of our experiment suggest that MLG #86 may have outcompeted other strains. Intraspecific functional diversity is still an unexplored topic for phytoplankton. However, the different susceptibility to parasite attacks by genetically distinct strains of the same diatom species (De Bruin et al., 2004) are examples that such differences exist in unicellular microalgae.
Conclusions
The results of our study on the planktonic marine diatom P. multistriata showed the alternation over relatively short temporal scales of abrupt clonal expansion of genetically related genotypes and the re-shuffling effect of sexual recombination. Moreover, strongly unbalanced mating type ratios were recorded during the contemporarily demographic and clonal expansion event that we recorded in 2013. These mechanisms and the characteristic population dynamics of unicellular microalgae, in which blooms are followed by severe bottlenecks between one bloom and the other, produce a nonlinear temporal pattern with variable levels of interannual differentiation. Long-term population genetic data sets and the availability of genomic resources (Basu et al., 2017) open the way to further studies that will shed light on the micro-evolutionary processes in this toxic diatom and their role in creating standing genetic variation upon which selection acts.
Acknowledgments
We thank Carmen Minucci for technical help in genotyping; Eleonora Scalco and Christophe Legrand for providing additional data on mating types; Diana Sarno for providing cell abundance data for P. multistriata; Augusto Passarelli for providing physical-chemical data at LTER-MC; the Molecular Biology and Bioinformatic Unit for sequencing and the Monitoring and Environmental Data Unit (MEDA) for sampling. The study was funded by Flagship Project RITMARE—The Italian Research for the Sea—coordinated by the Italian National Research Council and funded by the Italian Ministry of Education, University and Research within the National Research Program 2011–2013; LV was supported by a PhD fellowship from Stazione Zoologica Anton Dohrn (SZN).
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990). Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Arnaud-Haond S, Belkhir K. (2007). GENCLONE: a computer program to analyse genotypic data, test for clonality and describe spatial clonal organization. Mol Ecol Resour 7: 15–17. [Google Scholar]
- Basu S, Patil S, Mapleson D, Russo M, Vitale L, Fevola C et al. (2017). Finding a partner in the ocean: molecular and evolutionary bases of the response to sexual cues in a planktonic diatom. New Phytol 215: 140–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrenfeld MJ, Boss E, Siegel DA, Shea DM. (2005). Carbon-based ocean productivity and phytoplankton physiology from space. Global Biogeochem Cycles 19: GB1006. [Google Scholar]
- Bengtsson BO. (2003). Genetic variation in organisms with sexual and asexual reproduction. J Evol Biol 16: 189–199. [DOI] [PubMed] [Google Scholar]
- Berdalet E, Fleming LE, Gowen R, Davidson K, Hess P, Backer LC et al. (2016). Marine harmful algal blooms, human health and wellbeing: challenges and opportunities in the 21st century. J Mar Biol Assoc U K 96: 61–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alelio D, Amato A, WHCF Kooistra, Procaccini G, Casotti R, Montresor M. (2009. a). Internal transcribed spacer polymorphism in Pseudo-nitzschia multistriata (Bacillariophyceae) in the Gulf of Naples: recent divergence or intraspecific hybridization? Protist 160: 9–20. [DOI] [PubMed] [Google Scholar]
- D’Alelio D, Amato A, Luedeking A, Montresor M. (2009. b). Sexual and vegetative phases in the planktonic diatom Pseudo-nitzschia multistriata. Harmful Algae 8: 225–232. [Google Scholar]
- D’Alelio D, Gandolfi A. (2012). Recombination signals in the rpoC1 gene indicate gene-flow between Planktothrix (Cyanoprokaryota) species. J Phycol 48: 1424–1432. [DOI] [PubMed] [Google Scholar]
- D’Alelio D, Libralato S, Wyatt T, Ribera d’Alcalà M. (2016). Ecological-network models link diversity, structure and function in the plankton food-web. Sci Rep 6: 21806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alelio D, Ribera d’Alcalà M, Dubroca L, Sarno D, Zingone A, Montresor M. (2010). The time for sex: a biennial life cycle in a marine planktonic diatom. Limnol Oceanogr 55: 106–114. [Google Scholar]
- D’Alelio D, Ruggiero MV. (2015). Interspecific plastidial recombination in the diatom genus Pseudo-nitzschia. J Phycol 51: 1024–1028. [DOI] [PubMed] [Google Scholar]
- D’Alelio D, Salmaso N, Gandolfi A. (2013). Frequent recombination shapes the epidemic population structure of Planktothrix (Cyanoprokaryota) in Italian subalpine lakes. J Phycol 49: 1107–1117. [DOI] [PubMed] [Google Scholar]
- De Bruin A, Ibelings BW, Rijkeboer M, Brehm M, van Donk E. (2004). Genetic variation in Asterionella formosa (Bacillariophyceae): is it linked to frequent epidemics of host-specific parasitic fungi? J Phycol 40: 823–830. [Google Scholar]
- Dia A, Guillou L, Mauger S, Bigeard E, Marie D, Valero M et al. (2014). Spatiotemporal changes in the genetic diversity of harmful algal blooms caused by the toxic dinoflagellate Alexandrium minutum. Mol Ecol 23: 549–560. [DOI] [PubMed] [Google Scholar]
- Erdner DL, Richlen M, McCauley LAR, Anderson DM. (2011). Diversity and dynamics of a widespread bloom of the toxic dinoflagellate Alexandrium fundyense. PLoS One 6: e22965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godhe A, Kremp A, Montresor M. (2014). Genetic and microscopic evidence for sexual reproduction in the centric diatom Skeletonema marinoi. Protist 165: 401–416. [DOI] [PubMed] [Google Scholar]
- Godhe A, Sjöqvist C, Sildever S, Sefbom J, Harðardóttir S, Bertos-Fortis M et al. (2016). Physical barriers and environmental gradients cause spatial and temporal genetic differentiation of an extensive algal bloom. J Biogeogr 43: 1130–1142. [Google Scholar]
- Gsell AS, de Senerpont Domis LN, Verhoeven KJF, van Donk E, Ibelings BW. (2013). Chytrid epidemics may increase genetic diversity of a diatom spring-bloom. ISME J 7: 2057–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillard RRL. (1975). Culture of phytoplankton for feeding marine invertebrates. In: Culture of marine invertebrate animals. Springer, pp 29–60. [Google Scholar]
- Härnström K, Ellegaard M, Andersen TJ, Godhe A. (2011). Hundred years of genetic structure in a sediment revived diatom population. Proc Natl Acad Sci USA 108: 4252–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubold B, Hudson RR. (2000). LIAN 3.0: detecting linkage disequilibrium in multilocus data. Bioinformatics 16: 847. [DOI] [PubMed] [Google Scholar]
- Krueger-Hadfield SA, Balestreri C, Schroeder J, Highfield A, Helaouët P, Allum J et al. (2014). Genotyping an Emiliania huxleyi (prymnesiophyceae) bloom event in the North Sea reveals evidence of asexual reproduction. Biogeosciences 11: 5215–5234. [Google Scholar]
- MacLeod A, Tweedie A, Welburn SC, Maudlin I, Turner CMR, Tait A. (2000). Minisatellite marker analysis of Trypanosoma brucei: Reconciliation of clonal, panmictic, and epidemic population genetic structures. Proc Natl Acad Sci USA 97: 13442–13447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiden MCJ. (2006). Multilocus sequence typing of bacteria. Annu Rev Microbiol 60: 561–588. [DOI] [PubMed] [Google Scholar]
- McCabe RM, Hickey BM, Kudela RM, Lefebvre KA, Adams NG, Bill BD et al. (2016). An unprecedented coastwide toxic algal bloom linked to anomalous ocean conditions. Geophys Res Lett 43: 10,366–10,376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall ROD, Smouse PE. (2006). GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6: 288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton TJ, DeGiorgio M, Rosenberg NA. (2013). Population structure in a comprehensive genomic data set on human microsatellite variation. G3 3: 891–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond M, Rousset F. (1995). GENEPOP (Version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86: 248. [Google Scholar]
- Rengefors K, Kremp A, Reusch TBH, Wood AM. (2017). Genetic diversity and evolution in eukaryotic phytoplankton: revelations from population genetic studies. J Plankton Res 39: 165–179. [Google Scholar]
- Ribera d’Alcalà M, Conversano F, Corato F, Licandro P, Mangoni O, Marino D et al. (2004). Seasonal patterns in plankton communities in a pluriannual time series at a coastal Mediterranean site (Gulf of Naples): an attempt to discern recurrences and trends. Sci Mar 68: 65–83. [Google Scholar]
- Richlen ML, Erdner DL, McCauley LAR, Liberal K, Anderson DM. (2012). Extensive genetic diversity and rapid population differentiation during blooms of Alexandrium fundyense (dinophyceae) in an isolated salt pond on cape cod, MA, USA. Ecol Evol 2: 2588–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rynearson TA, Armbrust EV. (2005). Maintenance of clonal diversity during a spring bloom of the centric diatom Ditylum brightwellii. Mol Ecol 14: 1631–1640. [DOI] [PubMed] [Google Scholar]
- Saravanan V, Godhe A. (2010). Genetic heterogeneity and physiological variation among seasonally separated clones of Skeletonema marinoi (Bacillariophyceae) in the Gullmar Fjord, Sweden. Eur J Phycol 45: 177–190. [Google Scholar]
- Schurko AM, Logsdon JM, Eads BD. (2009. a). Meiosis genes in Daphnia pulex and the role of parthenogenesis in genome evolution. BMC Evol Biol 9: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurko AM, Neiman M, Logsdon JM. (2009. b). Signs of sex: what we know and how we know it. Trends Ecol Evol 24: 208–217. [DOI] [PubMed] [Google Scholar]
- Smayda TJ. (1997). What is a bloom? Limnol Oceanogr 42: 1132–1136. [Google Scholar]
- Smith JM, Smith NH, O’Rourke M, Spratt BG. (1993). How clonal are bacteria? Proc Natl Acad Sci USA 90: 4384–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speijer D, Lukeš J, Eliáš M. (2015). Sex is a ubiquitous, ancient, and inherent attribute of eukaryotic life. Proc Natl Acad Sci USA 112: 8827–8834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckel S, Masson JP. (2014). The exact distributions of FISunder partial asexuality in small finite populations with mutation. PLoS One 9: e85228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesson SVM, Borra M, Kooistra W, Procaccini G. (2011). Microsatellite primers in the planktonic diatom Pseudo-nitzschia multistriata (Bacillariophyceae). Am J Bot 98: E33–E35. [DOI] [PubMed] [Google Scholar]
- Tesson SVM, Legrand C, van Oosterhout C, Montresor M, Kooistra WHCF, Procaccini G. (2013). Mendelian inheritance pattern and high mutation rates of microsatellite alleles in the diatom Pseudo-nitzschia multistriata. Protist 164: 89–100. [DOI] [PubMed] [Google Scholar]
- Tesson SVM, Montresor M, Procaccini G, Kooistra WHCF. (2014). Temporal changes in population structure of a marine planktonic diatom. PLoS One 9: e114984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibayrenc M, Ayala FJ. (2012). Reprodaductive clonality of pathogens: a perspective on pathogenic viruses, bacteria, fungi, and parasitic protozoa. Proc Natl Acad Sci USA 109: E3305–E3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanormelingen P, Evans KM, Mann DG, Lance S, Debeer AE, D’Hondt S et al. (2015). Genotypic diversity and differentiation among populations of two benthic freshwater diatoms as revealed by microsatellites. Mol Ecol 24: 4433–4448. [DOI] [PubMed] [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. (2004). MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4: 535–538. [Google Scholar]
- von Dassow P, Montresor M. (2010). Unveiling the mysteries of phytoplankton life cycles: patterns and opportunities behind complexity. J Plankton Res 33: 3–12. [Google Scholar]
- Weedall GD, Hall N. (2015). Sexual reproduction and genetic exchange in parasitic protists. Parasitology 142: S120–S127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt T. (2012). Margalef’s mandala and phytoplankton bloom strategies. Deep Sea Res Part II Top Stud Oceanogr 101: 32–49. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.