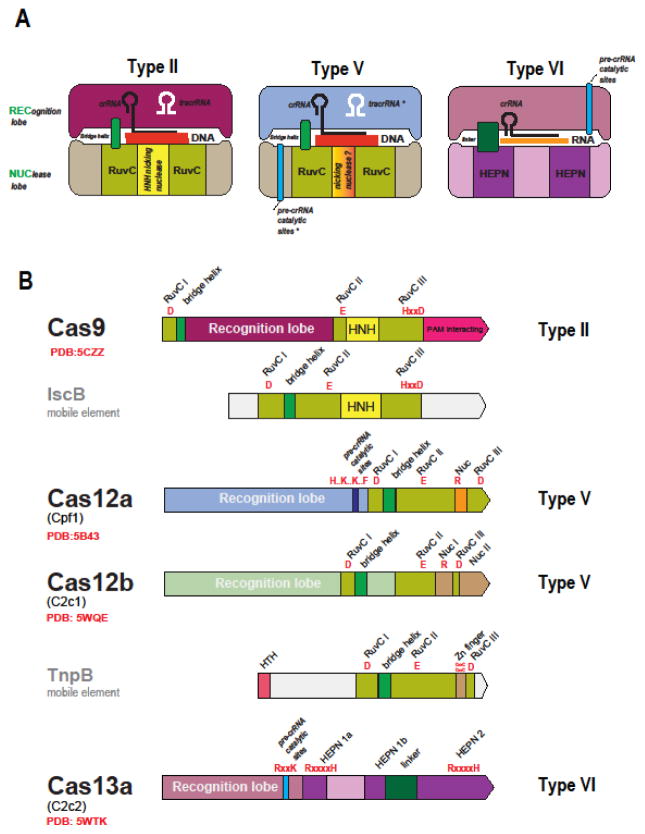
Figure 3. The Class 2 effector proteins.
(A) Schematic representation of the complexes of effector proteins with the target DNA or RNA, guide RNA and (for type II) tracrRNA
(B) Domain architectures of the effector proteins and their transposon-encoded evolutionary ancestors
IscB and TnpB are the inferred ancestors of the type II (Cas9) and type V (Cas12) effectors, respectively. The catalytic residues of the effector nuclease domain and, for Cas12a and Cas13a, the residues shown to be required for pre-crRNA processing are indicated in red. The Protein Data Bank (PDB) codes are included for proteins with solved structures. HTH, helix-turn-helix DNA-binding domain. The tracrRNA, the pre-crRNA processing catalytic sites and the nicking, target strand-cleaving nuclease are denoted by asterisks to indicate that they are each present only in subsets of the type II and type V effectors 9see text for details). The catalytic amino acid residues of the target-cleaving nucleases are shown in red, and those of the pre-crRNA processing nuclease are shown in blue. RuvCI, II and III stand for the three conserved motifs of the RuvC-like nuclease domain that contain the catalytic residues. In the motif signature, “x” stands for amino acid, and “..” indicated that the catalytic residues are separated by a small, variable number of non-conserved residues.