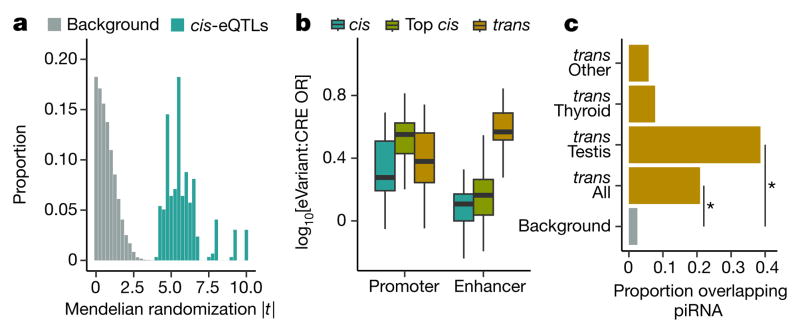
Figure 4. Functional characterization of GTEx trans-eVariants.
a, Frequency distribution of Mendelian randomization t-statistic, for 296 cis–trans-eQTLs and matched background variants. b, CRE enrichment (y-axis) of trans-eVariants (10% FDR), cis-eVariants (10% FDR, to match trans-eVariants), and top most significant cis-eVariants. Box plots show promoter and enhancer enrichment (x-axis) in matched cell-type CRE annotations compared to MAF- and distance-matched background variants. c, Proportion (x-axis) of variants overlapping piRNA clusters, including randomly sampled background loci, trans-eVariants across all tissues, testis trans-eVariants, thyroid trans-eVariants, and trans-eVariants from all tissues other than testis and thyroid. Asterisks denote significant enrichment (permutation test, P ≤1.0 ×10−4). Box plots depict the IQR, whiskers depict 1.5× IQR.