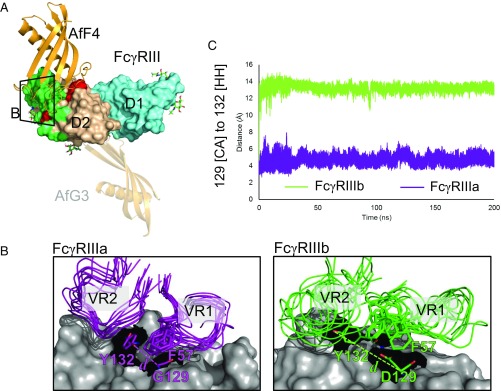
Fig. 3.
Molecular basis of interaction specificity of AfF4 for FcγRIIIa over FcγRIIIb. (A) Overview of AfF4 (orange) interaction with FcγRIIIa/b (domain 1 in aquamarine and domain 2 in wheat) showing FcγRIIIa/b-discriminating residues in red and IgG contacts in green. N-linked glycans with distinguishable electron density in the crystal structures are depicted as sticks and are listed in Table S6. The zoomed area for B is also shown. The AfG3-binding position is depicted as a transparent cartoon to aid comparison. (B) The AfF4 VR interface in FcγRIIIa and FcγRIIIb from MD simulations. AfF4 VR1 and VR2 are depicted as ensembles of snapshots taken at 20-ns intervals in representative simulations of the FcγRIIIa– and FcγRIIIb–AfF4 interactions. Interatomic distances between γa/b-Gly/Asp129 [CA] and γa/b-Tyr132 [HH] are illustrated as dashed lines and are represented in dynamic measurements in C. In FcγRIIIa the γa-Tyr132 sidechain orientates toward γa-Gly129, enabling AfF4 VR2 to form stable hydrophobic interactions involving γa-Tyr132 and AfF4-Phe57. In FcγRIIIb the γb-Asp129 sidechain clashes with γb-Tyr132, causing the ring to orientate away from γb-Asp129. γb-Asp129 also clashes with AfF4-Phe57, leading to disruption of the AfF4 VR2 hydrophobic interaction and higher mobility of VR2 and AfF4-Phe57 in FcγRIIIb. (C) Interatomic distance d averaged over triplicate MD simulations of FcγRIIIa– and FcγRIIIb–AfF4 interactions.