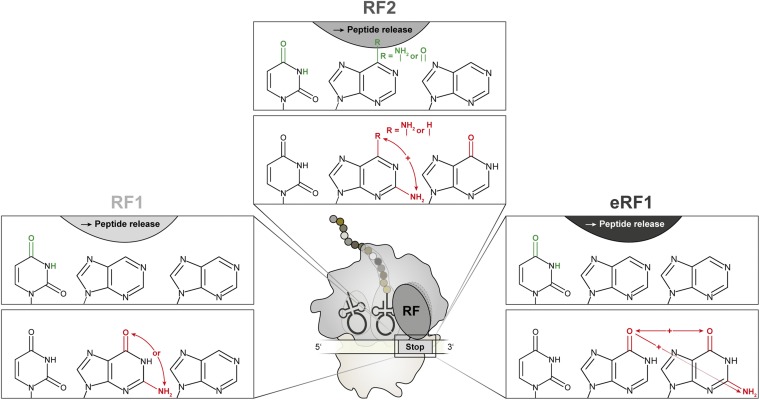
Fig. 5.
Summary of the chemical requirements of stop codon nucleotides for recognition by prokaryotic RF1 (Left), RF2 (Center), and eukaryotic eRF1 (Right). The pyrimidine–purine–purine matrix of all stop codons is schematically depicted (black). Exocyclic groups that were identified to be crucial for stop codon recognition and peptide release by RFs are illustrated in green (Upper). Chemical groups that were discriminated by the respective RF are shown in red (Lower). + indicates inhibitory effects mediated by a combination of distinct exocyclic groups of the stop codon.