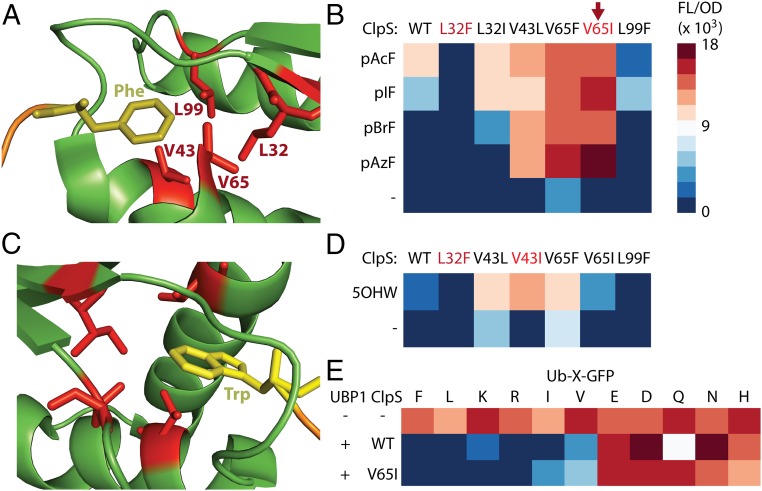
Fig. 2.
Proofreading tunability achieved through rational ClpS engineering. (A) Cartoon generated from crystal structure of E. coli ClpS binding N-end Phe peptide (PDB ID code: 3O2B) showing four hydrophobic ClpS residues subjected to single-point mutations that sampled F/L/I/V. (B) Heatmap of FL/OD signals obtained using a ClpS− host expressing UBP1, the p-acetyl-phenylalanine OTS, and variants of ClpS in the presence of different nsAAs. (C) Cartoon generated from crystal structure of C. crescentus ClpS binding N-end Trp peptide (PDB ID code: 3GQ1). (D) FL/OD heatmap resulting from expression of UBP1, the 5-hydroxytryptophan OTS, and ClpS variants in the presence/absence of 5-hydroxytryptophan. Scale as in B. (E) FL/OD heatmap resulting from expression of UBP1/ClpS in strains with Ub-X-GFP reporter genes expressing standard AAs in place of X.