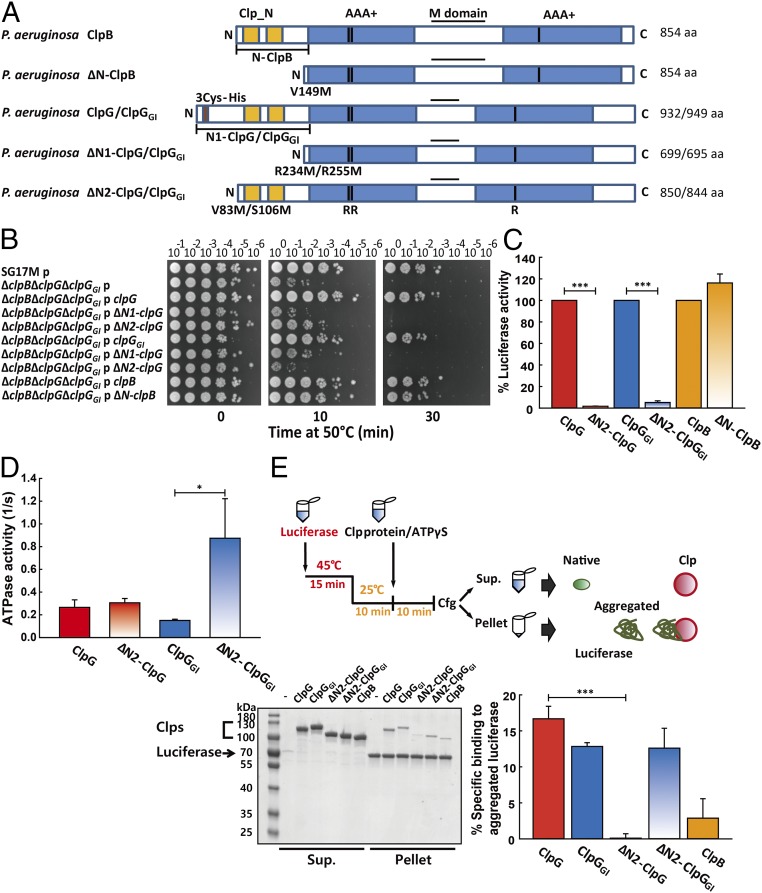
Fig. 6.
The unique N-domain extensions of ClpG/ClpGGI are essential for disaggregation activity. (A) Domain organization of ClpG/ClpGGI and ClpB. The generated N-terminal deletion variants of ClpG/ClpGGI (ΔN1 and ΔN2) and ClpB (ΔN) are indicated (B) P. aeruginosa SG17M ∆clpB ∆clpG ∆clpGGI expressing full-length, N-terminal truncated ClpG/ClpGGI or ClpB proteins from the vector pJN105 were subjected to heat shock at 50 °C for 10 min and 30 min. Cellular viabilities were determined by spotting serial dilutions (100–10−6) of cells on LB plates. p = pJN105. (C) Refolding of heat-aggregated luciferase was monitored in the presence of indicated chaperones. The activity of native luciferase in the presence of wild-type chaperones ClpG, ClpGGI, and ClpB after 90 min of refolding was set as 100%. (D) Basal ATPase activities of indicated ClpG/ClpGGI variants were determined. (E) Experimental scheme for monitoring binding of ClpG/ClpGGI and ClpB proteins to heat-aggregated luciferase. (Left) Heat-aggregated luciferase was incubated with the indicated chaperones in the presence of 2 mM ATPγS. Soluble (Sup.) and insoluble (pellet) fractions were analyzed by SDS-PAGE. (Right) Intensities of protein bands were quantified by ImageJ with the fraction (%) of chaperone present in the pellet fraction versus chaperone without luciferase aggregates calculated. Binding experiments were performed three times with a representative SDS-PAGE shown. Quantifications are based on three independent experiments (*P < 0.05; ***P < 0.001).