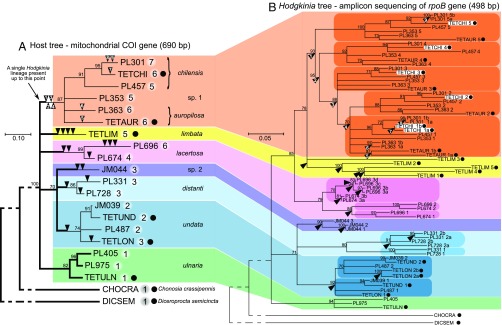
Fig. 2.
Maximum likelihood phylogenies of cicadas from the genus Tettigades (A) and of their Hodgkinia symbionts (B), based on partial sequences of mitochondrial cytochrome oxidase I (COI) gene and RNA polymerase subunit B (rpoB) gene, respectively. The phylogenies have been constrained based on multigene phylogenies for samples with genomes sequenced, which are indicated by black dots following leaf labels (SI Appendix, Fig. S2). Bootstrap values of 70% or more are shown as percentages above the nodes, and nodes with lower support have been collapsed. Colors represent species or species groups in which the ancestral state was a single lineage of Hodgkinia. In the host tree, thick branch lines indicate hosts, or the times in their evolutionary history when they hosted only a single Hodgkinia genotype, and numbers after leaf labels represent the number of distinct Hodgkinia rpoB genotypes in that sample, based on genomic data (when available) or rpoB amplicon data. Black arrowheads indicate Hodgkinia splitting events, as opposed to codiversification with hosts. The splits have been individually numbered in the chilensis-sp.1-auropilosa clade to more clearly show the inferred order of events. In the Hodgkinia tree, colored ovals identify clades that include strains from all sampled hosts in a species group, indicating that the first Hodgkinia split happened in the common ancestor of all sampled hosts from that clade. Note that Hodgkinia strains from a single cicada specimen occur in multiple places in the symbiont tree, as illustrated by TETCHI (shown inside white rectangles).