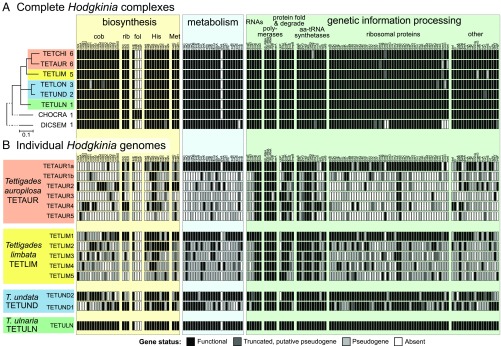
Fig. 4.
The list of all nonhypothetical protein-coding and RNA (other than tRNA) genes identified in the genomes of Hodgkinia from DICSEM, CHOCRA, and Tettigades spp. The classification (apparently functional, pseudogenized, or absent) of each gene is based on the length of the ORF relative to that in the genome of TETULN Hodgkinia. (A) At the level of a Hodgkinia complex, the highest level functional state of a gene in any of the genomes of a complex is provided. Host phylogeny is based on 15 mitochondrial genes (SI Appendix, Fig. S2), the number of distinct Hodgkinia lineages in a given specimen is provided after each leaf label, and host clades where Hodgkinia splits have happened independently are indicated with colored rectangles. (B) At the level of individual Hodgkinia genomes from four complexes, representing three independent splitting events. cob, cobalamin; fol, folate; rib, riboflavin.